BRN2 suppresses apoptosis, reprograms DNA damage repair, and is associated with a high somatic mutation burden in melanoma
- PMID: 30804224
- PMCID: PMC6411009
- DOI: 10.1101/gad.314633.118
BRN2 suppresses apoptosis, reprograms DNA damage repair, and is associated with a high somatic mutation burden in melanoma
Abstract
Whether cell types exposed to a high level of environmental insults possess cell type-specific prosurvival mechanisms or enhanced DNA damage repair capacity is not well understood. BRN2 is a tissue-restricted POU domain transcription factor implicated in neural development and several cancers. In melanoma, BRN2 plays a key role in promoting invasion and regulating proliferation. Here we found, surprisingly, that rather than interacting with transcription cofactors, BRN2 is instead associated with DNA damage response proteins and directly binds PARP1 and Ku70/Ku80. Rapid PARP1-dependent BRN2 association with sites of DNA damage facilitates recruitment of Ku80 and reprograms DNA damage repair by promoting Ku-dependent nonhomologous end-joining (NHEJ) at the expense of homologous recombination. BRN2 also suppresses an apoptosis-associated gene expression program to protect against UVB-, chemotherapy- and vemurafenib-induced apoptosis. Remarkably, BRN2 expression also correlates with a high single-nucleotide variation prevalence in human melanomas. By promoting error-prone DNA damage repair via NHEJ and suppressing apoptosis of damaged cells, our results suggest that BRN2 contributes to the generation of melanomas with a high mutation burden. Our findings highlight a novel role for a key transcription factor in reprogramming DNA damage repair and suggest that BRN2 may impact the response to DNA-damaging agents in BRN2-expressing cancers.
Keywords: BCL2; BRN2; Ku80; POU3F2; apoptosis; melanoma; nonhomologous end-joining; somatic mutation burden.
© 2019 Herbert et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


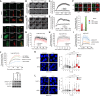


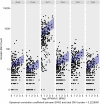

References
-
- Andersen B, Rosenfeld MG. 2001. POU domain factors in the neuroendocrine system: lessons from developmental biology provide insights into human disease. Endocr Rev 22: 2–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
