Synergistic lethality between BRCA1 and H3K9me2 loss reflects satellite derepression
- PMID: 30804228
- PMCID: PMC6446544
- DOI: 10.1101/gad.322495.118
Synergistic lethality between BRCA1 and H3K9me2 loss reflects satellite derepression
Abstract
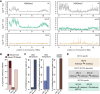
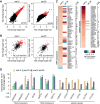
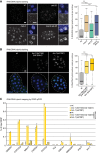
Caenorhabditis elegans has two histone H3 Lys9 methyltransferases, MET-2 (SETDB1 homolog) and SET-25 (G9a/SUV39H1 related). In worms, we found simple repeat sequences primarily marked by H3K9me2, while transposable elements and silent tissue-specific genes bear H3K9me3. RNA sequencing (RNA-seq) in histone methyltransferase (HMT) mutants shows that MET-2-mediated H3K9me2 is necessary for satellite repeat repression, while SET-25 silences a subset of transposable elements and tissue-specific genes through H3K9me3. A genome-wide synthetic lethality screen showed that RNA processing, nuclear RNA degradation, the BRCA1/BARD1 complex, and factors mediating replication stress survival are necessary for germline viability in worms lacking MET-2 but not SET-25. Unlike set-25 mutants, met-2-null worms accumulated satellite repeat transcripts, which form RNA:DNA hybrids on repetitive sequences, additively with the loss of BRCA1 or BARD1. BRCA1/BARD1-mediated H2A ubiquitination and MET-2 deposited H3K9me2 on satellite repeats are partially interdependent, suggesting both that the loss of silencing generates BRCA-recruiting DNA damage and that BRCA1 recruitment by damage helps silence repeats. The artificial induction of MSAT1 transcripts can itself trigger damage-induced germline lethality in a wild-type background, arguing that the synthetic sterility upon BRCA1/BARD1 and H3K9me2 loss is directly linked to the DNA damage provoked by unscheduled satellite repeat transcription.
Keywords: BRCA1 complex; DNA repeats; RNA:DNA hybrids; genome instability; heterochromatin; histone H3K9 methylation; satellite repeats; transcriptional silencing.
© 2019 Padeken et al.; Published by Cold Spring Harbor Laboratory Press.
Figures




References
-
- Avgustinova A, Symeonidi A, Castellanos A, Urdiroz-Urricelqui U, Solé-Boldo L, Martín M, Pérez-Rodríguez I, Prats N, Lehner B, Supek F, et al. 2018. Loss of G9a preserves mutation patterns but increases chromatin accessibility, genomic instability and aggressiveness in skin tumours. Nat Cell Biol 20: 1400–1409. 10.1038/s41556-018-0233-x - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
