USP7 Regulates Cytokinesis through FBXO38 and KIF20B
- PMID: 30804394
- PMCID: PMC6389929
- DOI: 10.1038/s41598-019-39368-y
USP7 Regulates Cytokinesis through FBXO38 and KIF20B
Abstract
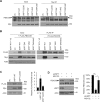
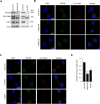
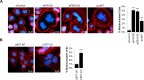
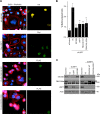
The ubiquitin specific protease 7 (USP7 or HAUSP) is known to regulate a variety of cellular processes by binding and deubiquitylating specific target proteins. To gain a more comprehensive understanding of its interactions and functions, we used affinity purification coupled to mass spectrometry to profile USP7 interactions. This revealed a novel interaction with FBXO38, a poorly characterized F-box protein. We showed that USP7 stabilizes FBXO38 dependent on its catalytic activity by protecting FBXO38 from proteasomal degradation. We used a BioID approach to profile the protein interactions (and putative functions) of FBXO38, revealing an interaction with KIF20B, a Kinesin-6 protein required for efficient cytokinesis. FBXO38 was shown to function independently from an SCF complex to stabilize KIF20B. Consequently, depletion of either FBXO38 or USP7 led to dramatic decreases in KIF20B levels and KIF20B at the midbody, which were manifested in cytokinetic defects. Furthermore, cytokinetic defects associated with USP7 silencing were rescued by restoring FBXO38 or KIF20B. The results indicate a novel mechanism of regulating cytokinesis through USP7 and FBXO38.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

