How peptide/MHC presence affects the dynamics of the LC13 T-cell receptor
- PMID: 30804417
- PMCID: PMC6389892
- DOI: 10.1038/s41598-019-38788-0
How peptide/MHC presence affects the dynamics of the LC13 T-cell receptor
Abstract
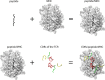
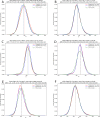
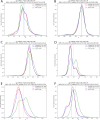
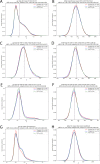
The interaction between T-cell receptors (TCRs) of T-cells and potentially immunogenic peptides presented by MHCs of antigen presenting cells is one of the most important mechanisms of the adaptive human immune system. A large number of structural simulations of the TCR/peptide/MHC system have been carried out. However, to date no study has investigated the differences of the dynamics between free TCRs and pMHC bound TCRs on a large scale. Here we present a study totalling 37 100 ns investigating the LC13 TCR in its free form as well as in complex with HLA-B*08:01 and different peptides. Our results show that the dynamics of the bound and unbound LC13 TCR differ significantly. This is reflected in (a) expected results such as an increased flexibility and increased solvent accessible surface of the CDRs of unbound TCR simulations but also in (b) less expected results such as lower CDR distances and compactness as well as alteration in the hydrogen bond network around CDR3α of unbound TCR simulations. Our study further emphasises the structural flexibility of TCRs and confirms the importance of the CDR3 loops for the adoption to MHC.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Kalyaanamoorthy S, Chen YP. Modelling and enhanced molecular dynamics to steer structure-based drug discovery. Prog. Biophys. Mol. Biol. 2014;114(3):123–136. - PubMed
-
- Sledz P, Caflisch A. Protein structure-based drug design: from docking to molecular dynamics. Curr. Opin. Struct. Biol. 2018;48:93–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

