Apical-basal polarity inhibits epithelial-mesenchymal transition and tumour metastasis by PAR-complex-mediated SNAI1 degradation
- PMID: 30804505
- PMCID: PMC6546105
- DOI: 10.1038/s41556-019-0291-8
Apical-basal polarity inhibits epithelial-mesenchymal transition and tumour metastasis by PAR-complex-mediated SNAI1 degradation
Erratum in
-
Author Correction: Apical-basal polarity inhibits epithelial-mesenchymal transition and tumour metastasis by PAR-complex-mediated SNAI1 degradation.Nat Cell Biol. 2023 Sep;25(9):1385. doi: 10.1038/s41556-023-01223-8. Nat Cell Biol. 2023. PMID: 37580389 No abstract available.
Abstract
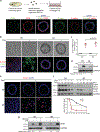
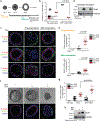
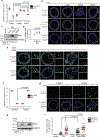
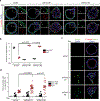
Loss of apical-basal polarity and activation of epithelial-mesenchymal transition (EMT) both contribute to carcinoma progression and metastasis. Here, we report that apical-basal polarity inhibits EMT to suppress metastatic dissemination. Using mouse and human epithelial three-dimensional organoid cultures, we show that the PAR-atypical protein kinase C (aPKC) polarity complex inhibits EMT and invasion by promoting degradation of the SNAIL family protein SNAI1. Under intact apical-basal polarity, aPKC kinases phosphorylate S249 of SNAI1, which leads to protein degradation. Loss of apical-basal polarity prevents aPKC-mediated SNAI1 phosphorylation and stabilizes the SNAI1 protein to promote EMT and invasion. In human breast tumour xenografts, inhibition of the PAR-complex-mediated SNAI1 degradation mechanism promotes tumour invasion and metastasis. Analyses of human breast tissue samples reveal negative correlations between PAR3 and SNAI1 protein levels. Our results demonstrate that apical-basal polarity functions as a critical checkpoint of EMT to precisely control epithelial-mesenchymal plasticity during tumour metastasis.
Conflict of interest statement
COMPETING FINANCIAL INTERESTS
The authors declare no financial and non-financial competing interests.
Figures


Comment in
-
Epithelial polarity limits EMT.Nat Cell Biol. 2019 Mar;21(3):299-300. doi: 10.1038/s41556-019-0284-7. Nat Cell Biol. 2019. PMID: 30824839 No abstract available.
Similar articles
-
Shp2 promotes metastasis of prostate cancer by attenuating the PAR3/PAR6/aPKC polarity protein complex and enhancing epithelial-to-mesenchymal transition.Oncogene. 2016 Mar 10;35(10):1271-82. doi: 10.1038/onc.2015.184. Epub 2015 Jun 8. Oncogene. 2016. PMID: 26050620
-
Control of the Epithelial-to-Mesenchymal Transition and Cancer Metastasis by Autophagy-Dependent SNAI1 Degradation.Cells. 2019 Feb 6;8(2):129. doi: 10.3390/cells8020129. Cells. 2019. PMID: 30736337 Free PMC article.
-
Elevated expression of Par3 promotes prostate cancer metastasis by forming a Par3/aPKC/KIBRA complex and inactivating the hippo pathway.J Exp Clin Cancer Res. 2017 Oct 10;36(1):139. doi: 10.1186/s13046-017-0609-y. J Exp Clin Cancer Res. 2017. PMID: 29017577 Free PMC article.
-
Epigenetic Regulation and Post-Translational Modifications of SNAI1 in Cancer Metastasis.Int J Mol Sci. 2021 Oct 14;22(20):11062. doi: 10.3390/ijms222011062. Int J Mol Sci. 2021. PMID: 34681726 Free PMC article. Review.
-
The Par3/Par6/aPKC complex and epithelial cell polarity.Exp Cell Res. 2013 Jun 10;319(10):1357-64. doi: 10.1016/j.yexcr.2013.03.021. Epub 2013 Mar 25. Exp Cell Res. 2013. PMID: 23535009 Review.
Cited by
-
SCRIB Promotes Proliferation and Metastasis by Targeting Hippo/YAP Signalling in Colorectal Cancer.Front Cell Dev Biol. 2021 Apr 15;9:656359. doi: 10.3389/fcell.2021.656359. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33937255 Free PMC article.
-
Actomyosin fibers DApPLE epithelial apical junctions.J Cell Biol. 2022 May 2;221(5):e202203035. doi: 10.1083/jcb.202203035. Epub 2022 Apr 20. J Cell Biol. 2022. PMID: 35442398 Free PMC article.
-
Snail family transcriptional repressor 1 radiosensitizes esophageal cancer via epithelial-mesenchymal transition signaling: From bioinformatics to integrated study.World J Gastrointest Oncol. 2025 Apr 15;17(4):97644. doi: 10.4251/wjgo.v17.i4.97644. World J Gastrointest Oncol. 2025. PMID: 40235866 Free PMC article.
-
Modelling Cancer Pathophysiology: Mechanisms and Changes in the Extracellular Matrix During Cancer Initiation and Early Tumour Growth.Cancers (Basel). 2025 May 15;17(10):1675. doi: 10.3390/cancers17101675. Cancers (Basel). 2025. PMID: 40427172 Free PMC article. Review.
-
Possible prognostic impact of PKCι genetic variants in prostate cancer.Cancer Cell Int. 2024 Jan 10;24(1):24. doi: 10.1186/s12935-023-03182-4. Cancer Cell Int. 2024. PMID: 38200472 Free PMC article.
References
-
- Zen K et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene 28, 2910–2918 (2009). - PubMed
-
- Assemat E, Bazellieres E, Pallesi-Pocachard E, Le Bivic A & Massey-Harroche D Polarity complex proteins. Biochim Biophys Acta 1778, 614–630 (2008). - PubMed
-
- Lee M & Vasioukhin V Cell polarity and cancer--cell and tissue polarity as a non-canonical tumor suppressor. J Cell Sci 121, 1141–1150 (2008). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

