Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host
- PMID: 30804542
- PMCID: PMC7096966
- DOI: 10.1038/s41564-019-0371-3
Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host
Abstract
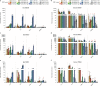
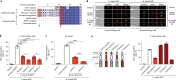
Bats are special in their ability to host emerging viruses. As the only flying mammal, bats endure high metabolic rates yet exhibit elongated lifespans. It is currently unclear whether these unique features are interlinked. The important inflammasome sensor, NLR family pyrin domain containing 3 (NLRP3), has been linked to both viral-induced and age-related inflammation. Here, we report significantly dampened activation of the NLRP3 inflammasome in bat primary immune cells compared to human or mouse counterparts. Lower induction of apoptosis-associated speck-like protein containing a CARD (ASC) speck formation and secretion of interleukin-1β in response to both 'sterile' stimuli and infection with multiple zoonotic viruses including influenza A virus (-single-stranded (ss) RNA), Melaka virus (PRV3M, double-stranded RNA) and Middle East respiratory syndrome coronavirus (+ssRNA) was observed. Importantly, this reduction of inflammation had no impact on the overall viral loads. We identified dampened transcriptional priming, a novel splice variant and an altered leucine-rich repeat domain of bat NLRP3 as the cause. Our results elucidate an important mechanism through which bats dampen inflammation with implications for longevity and unique viral reservoir status.
Conflict of interest statement
The authors declare no competing interests.
Figures



Comment in
-
"On the bat's back I do fly".Nat Rev Microbiol. 2019 May;17(5):265. doi: 10.1038/s41579-019-0177-6. Nat Rev Microbiol. 2019. PMID: 30842614 Free PMC article.
-
Bat tolerance to viral infections.Nat Microbiol. 2019 May;4(5):728-729. doi: 10.1038/s41564-019-0430-9. Nat Microbiol. 2019. PMID: 31015739 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

