Mutations in gyrB play an important role in ciprofloxacin-resistant Pseudomonas aeruginosa
- PMID: 30804676
- PMCID: PMC6371945
- DOI: 10.2147/IDR.S182272
Mutations in gyrB play an important role in ciprofloxacin-resistant Pseudomonas aeruginosa
Abstract
Purpose: To investigate the main molecular resistance mechanisms to fluoroquinolones (FQs) in Pseudomonas aeruginosa and also to investigate the effect of time and concentration on mutations in resistance genes.
Materials and methods: The clinical isolates of P. aeruginosa which are sensitive to ciprofloxacin (CIP) or levofloxacin (LEV) were collected. The isolates were incubated with different concentrations of CIP or LEV for 5 days and the minimal inhibitory concentrations (MICs) of CIP, LEV and ofloxacin (OFX) were measured. The MIC of FQs to P. aeruginosa was measured by the agar dilution method. FQ resistance determining regions of gyrA, gyrB, parC and parE were amplified by PCR, and mutations in four genes were explored using sequence analysis with the Snapgene software. The relative expression levels of two efflux pumps genes (mexA and mexE) were measured by quantitative reverse transcription PCR.
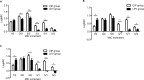
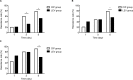
Results: A total of eleven isolates were collected from the Second Hospital of Shanxi Medical University. Amino acid alterations in gyrA and gyrB were mainly detected in resistant mutants, and the percentage of strains with amino acid alterations in gyrB was significantly higher than that in gyrA (P<0.001). MICs of strains with mutations both in gyrA and gyrB were not significantly higher than those of strains with mutations only in gyrB (P>0.05). No amino acid alterations were detected in genes of parC and parE. In both gyrA and gyrB, the number of amino acid alterations increased with incubation time prolonged and increased with increasing incubation concentration.
Conclusion: CIP was more competent than LEV in making P. aeruginosa resistant to in vitro selection. Mutations occurring in gyrB played an important role in FQ resistance of P. aeruginosa in vitro selection.
Keywords: Pseudomonas aeruginosa; fluoroquinolones; in vitro; molecular resistance mechanisms.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures


Similar articles
-
Mutations in the gyrA, parC, and mexR genes provide functional insights into the fluoroquinolone-resistant Pseudomonas aeruginosa isolated in Vietnam.Infect Drug Resist. 2018 Feb 28;11:275-282. doi: 10.2147/IDR.S147581. eCollection 2018. Infect Drug Resist. 2018. PMID: 29535543 Free PMC article.
-
[Susceptibility of 570 Pseudomonas aeruginosa strains to 11 antimicrobial agents and the mechanism of its resistance to fluoroquinolones].Zhonghua Yi Xue Za Zhi. 2003 Mar 10;83(5):403-7. Zhonghua Yi Xue Za Zhi. 2003. PMID: 12820918 Chinese.
-
The role of gyrA and parC mutations in fluoroquinolones-resistant Pseudomonas aeruginosa isolates from Iran.Braz J Microbiol. 2016 Oct-Dec;47(4):925-930. doi: 10.1016/j.bjm.2016.07.016. Epub 2016 Jul 26. Braz J Microbiol. 2016. PMID: 27522930 Free PMC article.
-
Deficiency of quorum sensing system inhibits the resistance selection of Pseudomonas aeruginosa to ciprofloxacin and levofloxacin in vitro.J Glob Antimicrob Resist. 2017 Sep;10:113-119. doi: 10.1016/j.jgar.2017.04.008. Epub 2017 Jul 17. J Glob Antimicrob Resist. 2017. PMID: 28729210
-
Type II topoisomerase mutations in ciprofloxacin-resistant strains of Pseudomonas aeruginosa.Antimicrob Agents Chemother. 1999 Jan;43(1):62-6. doi: 10.1128/AAC.43.1.62. Antimicrob Agents Chemother. 1999. PMID: 9869566 Free PMC article.
Cited by
-
Molecular Analysis of Pseudomonas aeruginosa Isolates with Mutant gyrA Gene and Development of a New Ciprofloxacin Derivative for Antimicrobial Therapy.Mol Biotechnol. 2025 Feb;67(2):649-660. doi: 10.1007/s12033-024-01076-y. Epub 2024 Feb 1. Mol Biotechnol. 2025. PMID: 38302682
-
Potential Synergistic Antibiotic Combinations against Fluoroquinolone-Resistant Pseudomonas aeruginosa.Pharmaceuticals (Basel). 2022 Feb 18;15(2):243. doi: 10.3390/ph15020243. Pharmaceuticals (Basel). 2022. PMID: 35215357 Free PMC article.
-
Rapid Phenotypic Convergence towards Collateral Sensitivity in Clinical Isolates of Pseudomonas aeruginosa Presenting Different Genomic Backgrounds.Microbiol Spectr. 2023 Feb 14;11(1):e0227622. doi: 10.1128/spectrum.02276-22. Epub 2022 Dec 19. Microbiol Spectr. 2023. PMID: 36533961 Free PMC article.
-
Detection of Pseudomonas aeruginosa pus wound isolate using a polymerase chain reaction targeting 16S rRNA and gyrB genes: A case from Indonesia.Narra J. 2024 Aug;4(2):e774. doi: 10.52225/narra.v4i2.774. Epub 2024 Jul 4. Narra J. 2024. PMID: 39280309 Free PMC article.
-
Novel fabrication of antibiotic containing multifunctional silk fibroin injectable hydrogel dressing to enhance bactericidal action and wound healing efficiency on burn wound: In vitro and in vivo evaluations.Int Wound J. 2022 Mar;19(3):679-691. doi: 10.1111/iwj.13665. Epub 2021 Aug 20. Int Wound J. 2022. PMID: 34414663 Free PMC article.
References
-
- Weiner LM, Webb AK, Limbago B, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the National Healthcare Safety Network at the Centers for Disease Control and Prevention, 2011–2014. Infect Control Hosp Epidemiol. 2016;37(11):1288–1301. - PMC - PubMed
-
- Nguyen L, Garcia J, Gruenberg K, Macdougall C. Multidrug-resistant Pseudomonas infections: hard to treat, but hope on the horizon? Curr Infect Dis Rep. 2018;20(8):23. - PubMed
-
- Qi Z, Duan M, Li A. Research status of drug resistance mechanism of Pseudomonas aeruginosa. Shandong Med J. 2014;54(4):83–86.
LinkOut - more resources
Full Text Sources

