Deep Neural Network Models for Predicting Chemically Induced Liver Toxicity Endpoints From Transcriptomic Responses
- PMID: 30804783
- PMCID: PMC6370634
- DOI: 10.3389/fphar.2019.00042
Deep Neural Network Models for Predicting Chemically Induced Liver Toxicity Endpoints From Transcriptomic Responses
Abstract
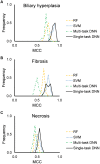
Improving the accuracy of toxicity prediction models for liver injuries is a key element in evaluating the safety of drugs and chemicals. Mechanism-based information derived from expression (transcriptomic) data, in combination with machine-learning methods, promises to improve the accuracy and robustness of current toxicity prediction models. Deep neural networks (DNNs) have the advantage of automatically assembling the relevant features from a large number of input features. This makes them especially suitable for modeling transcriptomic data, which typically contain thousands of features. Here, we gaged gene- and pathway-level feature selection schemes using single- and multi-task DNN approaches in predicting chemically induced liver injuries (biliary hyperplasia, fibrosis, and necrosis) from whole-genome DNA microarray data. The single-task DNN models showed high predictive accuracy and endpoint specificity, with Matthews correlation coefficients for the three endpoints on 10-fold cross validation ranging from 0.56 to 0.89, with an average of 0.74 in the best feature sets. The DNN models outperformed Random Forest models in cross validation and showed better performance than Support Vector Machine models when tested in the external validation datasets. In the cross validation studies, the effect of the feature selection scheme was negligible among the studied feature sets. Further evaluation of the models on their ability to predict the injury phenotype per se for non-chemically induced injuries revealed the robust performance of the DNN models across these additional external testing datasets. Thus, the DNN models learned features specific to the injury phenotype contained in the gene expression data.
Keywords: artificial neural network; biliary hyperplasia; classification model; liver fibrosis; liver necrosis; machine leaning; toxicity prediction.
Figures



References
-
- Breiman L. (2001). Random forests. Mach. Learn. 45 5–32. 10.1023/a:1010933404324 - DOI
LinkOut - more resources
Full Text Sources

