Identification of New HIV-1 Circulating Recombinant Forms CRF81_cpx and CRF99_BF1 in Central Western Brazil and of Unique BF1 Recombinant Forms
- PMID: 30804902
- PMCID: PMC6378278
- DOI: 10.3389/fmicb.2019.00097
Identification of New HIV-1 Circulating Recombinant Forms CRF81_cpx and CRF99_BF1 in Central Western Brazil and of Unique BF1 Recombinant Forms
Abstract
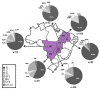
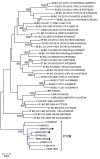
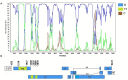
Intersubtype recombinants classified as circulating recombinant forms (CRFs) or unique recombinant forms (URFs) have been shown to play an important role in the complex and dynamic Brazilian HIV/AIDS epidemic. Previous pol region studies (2003-2013) in 828 patients from six states from Central Western, Northern and Northeastern Brazil reported variable rates of BF1, F1CB, BC, and CF1 mosaics. In this study HIV-1 subtype diversity BF1, F1CB, BC, and CF1 recombinants in pol were analyzed. Full/near-full/partial genome sequences were generated from F1CB and BF1 recombinants. Genomic DNA extracted from whole blood was used in nested-PCR to amplify four overlapping fragments encompassing the full HIV-1 genome. Phylogenetic trees were generated using the neighbor-joining/NJ method (MEGA 6.0). The time of the most recent common ancestor (TMRCA) of F1CB and BF1 clades was estimated using a Bayesian Markov Chain Monte Carlo approach (BEAST v1.8; BEAGLE). Bootscanning was used for recombination analyses (Simplot v3.5.1); separate NJ phylogenetic analysis of fragments confirmed subtypes. The phylogenetic analyses of protease/reverse-transcriptase sequences in 828 patients revealed 76% subtype B (n = 629), 6.4% subtype C (n = 53), 4.2% subtype F1 (n = 35), 13.4% intersubtype recombinants: 10.5% BF1 (n = 87), 2.3% BC (n = 19), 0.4% F1CB (n = 3), and 0.2% CF1 (n = 2). Two full and one partial BF1C genomes allowed the characterization of the CRF81_cpx that has 9 breakpoints dividing the genome into 10 subregions. Basic Local Alignment Search Tool searches (Los Alamos HIV Sequence Database) identified six other sequences with the same recombination profile in pol, five from Brazil, and one from Italy. The estimated median TMRCA of CRF81_cpx was 1999 (1992-2003). CRF60_BC-like sequences, originally described in Italy, were also found. Two full and one near full-length BF1 genomes led to the characterization of the new CRF99_BF1 that has six recombination breakpoints dividing the genome into seven subregions. Two new URFs BF1, with six recombination breakpoints and seven subregions were also characterized. The description of the first Brazilian BF1C CRF81_cpx and of the new CRF99_BF1 corroborate the important role of CRFs in the HIV/AIDS epidemic throughout Brazil. Our data also highlight the value of HIV-1 full-genome sequence studies in order to fully reveal the complexity of the epidemic in a huge country as Brazil.
Keywords: Brazil; CRFs; Central Western; HIV-1; URFs; molecular epidemiology.
Figures






References
-
- Alcântara K. C., Lins J. B., Albuquerque M., Aires L. M., Cardoso L. P., Minuzzi A. L., et al. (2012). HIV-1 mother-to-child transmission and drug resistance among Brazilian pregnant with high access to diagnosis and prophylactic measures. J. Clin. Virol. 54 15–20. 10.1016/j.jcv.2012.01.011 - DOI - PubMed
-
- Barreto C. C., Nishyia A., Araujo L. V., Ferreira J. E., Busch M. P., Sabino E. C., et al. (2006). Trends in antiretroviral drug resistance and clade distributions among HIV-1infected blood donors in Sao Paulo. Brazil. J. Acquir. Immune. Defic. Syndr. 41 338–341. 10.1097/01.qai.0000199097.88344.50 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

