Positive Regulation of Spoilage Potential and Biofilm Formation in Shewanella baltica OS155 via Quorum Sensing System Composed of DKP and Orphan LuxRs
- PMID: 30804914
- PMCID: PMC6370745
- DOI: 10.3389/fmicb.2019.00135
Positive Regulation of Spoilage Potential and Biofilm Formation in Shewanella baltica OS155 via Quorum Sensing System Composed of DKP and Orphan LuxRs
Abstract
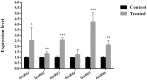
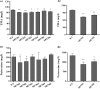
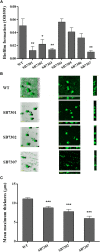
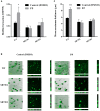
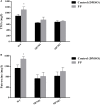
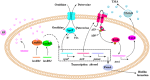
The spoilage potential and biofilm formation of Shewanella baltica are reported to be regulated by Quorum sensing (QS) system from the phenotype point of view, but the specific mechanism is not fully understood. In the present study, the QS autoinducers were detected by UHPLC-MS/MS, cell density-dependent luxR-type genes were obtained through autoregulation experiments among a series of candidates in S. baltica OS155 (The SSO of large yellow croaker). The direct interaction between cyclo-(L-Pro-L-Phe) (PP) and LuxR01 as well as LuxR02 proteins was revealed via in vitro binding assay. Deletion of luxR-type genes (luxR01 and luxR02) impaired spoilage potential and biofilm formation of S. baltica OS155 in various degrees. Transcriptional analysis and qRT-PCR validation showed that spoilage and biofilm-related genes torS, speF, and pomA were down-regulated in luxR01 and luxR02 deletion strains. In addition, exogenous PP promoted spoilage potential and biofilm formation, which could be attenuated by luxR01 or luxR02 deletion. Our results revealed an explicit QS system employing PP as autoinducer and two orphan LuxRs as receptors which positively regulated spoilage capacity and biofilm formation via transcriptional regulation of corresponding genes in S. baltica OS155, which provides potential specific targets for seafood preservation involving QS system.
Keywords: biofilm formation; diketopiperazines; orphan LuxR-type proteins; quorum sensing; spoilage.
Figures

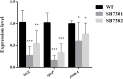
Similar articles
-
Quorum sensing signals affect spoilage of refrigerated large yellow croaker (Pseudosciaena crocea) by Shewanella baltica.Int J Food Microbiol. 2016 Jan 18;217:146-55. doi: 10.1016/j.ijfoodmicro.2015.10.020. Epub 2015 Oct 23. Int J Food Microbiol. 2016. PMID: 26519730
-
Inhibition of biofilm development and spoilage potential of Shewanella baltica by quorum sensing signal in cell-free supernatant from Pseudomonas fluorescens.Int J Food Microbiol. 2016 Aug 2;230:73-80. doi: 10.1016/j.ijfoodmicro.2016.04.015. Epub 2016 Apr 14. Int J Food Microbiol. 2016. PMID: 27149651
-
Inhibition of quorum sensing, biofilm, and spoilage potential in Shewanella baltica by green tea polyphenols.J Microbiol. 2015 Dec;53(12):829-36. doi: 10.1007/s12275-015-5123-3. Epub 2015 Dec 2. J Microbiol. 2015. PMID: 26626353
-
Quorum sensing in food spoilage and natural-based strategies for its inhibition.Food Res Int. 2020 Jan;127:108754. doi: 10.1016/j.foodres.2019.108754. Epub 2019 Oct 31. Food Res Int. 2020. PMID: 31882100 Review.
-
Exploring the Function of Quorum Sensing Regulated Biofilms in Biological Wastewater Treatment: A Review.Int J Mol Sci. 2022 Aug 28;23(17):9751. doi: 10.3390/ijms23179751. Int J Mol Sci. 2022. PMID: 36077148 Free PMC article. Review.
Cited by
-
Ornithine capture by a translating ribosome controls bacterial polyamine synthesis.Nat Microbiol. 2020 Apr;5(4):554-561. doi: 10.1038/s41564-020-0669-1. Epub 2020 Feb 24. Nat Microbiol. 2020. PMID: 32094585 Free PMC article.
-
Genomic Analysis of Two Representative Strains of Shewanella putrefaciens Isolated from Bigeye Tuna: Biofilm and Spoilage-Associated Behavior.Foods. 2022 Apr 27;11(9):1261. doi: 10.3390/foods11091261. Foods. 2022. PMID: 35563985 Free PMC article.
-
ARTP mutagenesis for genome-wide identification of genes important for biofilm regulation in spoilage bacterium Pseudomonas fluorescens PF08.Appl Environ Microbiol. 2025 Jun 18;91(6):e0021825. doi: 10.1128/aem.00218-25. Epub 2025 May 7. Appl Environ Microbiol. 2025. PMID: 40530873 Free PMC article.
-
Comparative Proteomics Reveals the Spoilage-Related Factors of Shewanella putrefaciens Under Refrigerated Condition.Front Microbiol. 2021 Dec 3;12:740482. doi: 10.3389/fmicb.2021.740482. eCollection 2021. Front Microbiol. 2021. PMID: 34925259 Free PMC article.
-
Novel Plasmid Carrying Mobile Colistin Resistance Gene mcr-4.3 and Mercury Resistance Genes in Shewanella baltica: Insights into Mobilization of mcr-4.3 in Shewanella Species.Microbiol Spectr. 2022 Dec 21;10(6):e0203722. doi: 10.1128/spectrum.02037-22. Epub 2022 Nov 14. Microbiol Spectr. 2022. PMID: 36374025 Free PMC article.
References
-
- Bai A. J., Rai Vittal R. (2014). Quorum sensing regulation and inhibition of exoenzyme production and biofilm formation in the food spoilage bacteria Pseudomonas psychrophila PSPF19. Food Biotechnol. 28 293–308. 10.1080/08905436.2014.963601 - DOI
-
- Baraquet C., Théraulaz L., Guiral M., Lafitte D., Méjean V., Jourlin-Castelli C. (2006). TorT, a member of a new periplasmic binding protein family, triggers induction of the tor respiratory system upon trimethylamineN-oxide electron-acceptor binding in Escherichia coli. J. Biol. Chem. 281 38189–38199. 10.1074/jbc.M604321200 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

