Semen Microbiome Biogeography: An Analysis Based on a Chinese Population Study
- PMID: 30804923
- PMCID: PMC6371047
- DOI: 10.3389/fmicb.2018.03333
Semen Microbiome Biogeography: An Analysis Based on a Chinese Population Study
Abstract
Investigating inter-subject heterogeneity (or spatial distribution) of human semen microbiome diversity is of important significance. Theoretically, the spatial distribution of biodiversity constitutes the core of microbiome biogeography. Practically, the inter-subject heterogeneity is crucial for understanding the normal (healthy) flora of semen microbiotas as well as their possible changes associated with abnormal fertility. In this article, we analyze the scaling (changes) of semen microbiome diversity across individuals with DAR (diversity-area relationship) analysis, a recent extension to classic SAR (species-area relationship) law in biogeography and ecology. Specifically, the unit of "area" is individual subject, and the microbial diversity in seminal fluid of an individual (area) is assessed via metagenomic DNA sequencing technique and measured in the Hill numbers. The DAR models were then fitted to the accrued diversity across different number of individuals (area size). We further tested the difference in DAR parameters among the healthy, subnormal, and abnormal microbiome samples in terms of their fertility status based on a cross-sectional study of a Chinese cohort. Given that no statistically significant differences in the DAR parameters were detected among the three groups, we built unified DAR models for semen microbiome by combining the healthy, subnormal, and abnormal groups. The model parameters were used to (i) estimate the microbiome diversity scaling in a population (cohort), and construct the so-termed DAR profile; (ii) predict/construct the maximal accrual diversity (MAD) profile in a population; (iii) estimate the pair-wise diversity overlap (PDO) between two individuals and construct the PDO profile; (iv) assess the ratio of individual diversity to population (RIP) accrual diversity. The last item (RIP) is a new concept we propose in this study, which is essentially a ratio of local diversity to regional or global diversity (LRD/LGD), applicable to general biodiversity investigation beyond human microbiome.
Keywords: DAR (diversity-area relationship); LRD/LGD (ratio of local to regional/global diversity); RIP (ratio of individual to population accrual diversity); beta-diversity; biogeography; inter-subject heterogeneity; semen microbiome.
Copyright © 2019 Ma and Li.
Figures
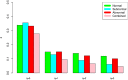


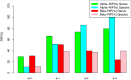
Similar articles
-
Global Microbiome Diversity Scaling in Hot Springs With DAR (Diversity-Area Relationship) Profiles.Front Microbiol. 2019 Feb 22;10:118. doi: 10.3389/fmicb.2019.00118. eCollection 2019. Front Microbiol. 2019. PMID: 30853941 Free PMC article.
-
Sketching the Human Microbiome Biogeography with DAR (Diversity-Area Relationship) Profiles.Microb Ecol. 2019 Apr;77(3):821-838. doi: 10.1007/s00248-018-1245-6. Epub 2018 Aug 28. Microb Ecol. 2019. PMID: 30155556
-
DAR (diversity-area relationship): Extending classic SAR (species-area relationship) for biodiversity and biogeography analyses.Ecol Evol. 2018 Sep 25;8(20):10023-10038. doi: 10.1002/ece3.4425. eCollection 2018 Oct. Ecol Evol. 2018. PMID: 30397444 Free PMC article.
-
The microbiome of the infertile male.Curr Opin Urol. 2020 May;30(3):355-362. doi: 10.1097/MOU.0000000000000742. Curr Opin Urol. 2020. PMID: 32235279 Review.
-
The emergence and promise of functional biogeography.Proc Natl Acad Sci U S A. 2014 Sep 23;111(38):13690-6. doi: 10.1073/pnas.1415442111. Epub 2014 Sep 15. Proc Natl Acad Sci U S A. 2014. PMID: 25225414 Free PMC article. Review.
Cited by
-
Diversity scaling of human vaginal microbial communities.Zool Res. 2019 Nov 18;40(6):587-594. doi: 10.24272/j.issn.2095-8137.2019.068. Zool Res. 2019. PMID: 31592582 Free PMC article.
-
Body fluids should be identified before estimating the time since deposition (TsD) in microbiome-based stain analyses for forensics.Microbiol Spectr. 2024 Apr 2;12(4):e0248023. doi: 10.1128/spectrum.02480-23. Epub 2024 Mar 12. Microbiol Spectr. 2024. PMID: 38470485 Free PMC article.
-
Semen dysbiosis-just a male problem?Front Cell Infect Microbiol. 2022 Sep 13;12:815786. doi: 10.3389/fcimb.2022.815786. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36176582 Free PMC article. Review.
-
Truffle species strongly shape their surrounding soil mycobiota in a Pinus armandii forest.Arch Microbiol. 2021 Dec;203(10):6303-6314. doi: 10.1007/s00203-021-02598-8. Epub 2021 Oct 15. Arch Microbiol. 2021. PMID: 34652507
-
Global Microbiome Diversity Scaling in Hot Springs With DAR (Diversity-Area Relationship) Profiles.Front Microbiol. 2019 Feb 22;10:118. doi: 10.3389/fmicb.2019.00118. eCollection 2019. Front Microbiol. 2019. PMID: 30853941 Free PMC article.
References
-
- Chao A., Chiu C. H., Jost L. (2014). Unifying species diversity, phylogenetic diversity, functional diversity and related similarity and differentiation measures through Hill numbers. Annu. Rev. Ecol. Evol. Syst. 45, 297–324. 10.1146/annurev-ecolsys-120213-091540 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

