Comparative Study of Immune Reaction Against Bacterial Infection From Transcriptome Analysis
- PMID: 30804945
- PMCID: PMC6370674
- DOI: 10.3389/fimmu.2019.00153
Comparative Study of Immune Reaction Against Bacterial Infection From Transcriptome Analysis
Abstract
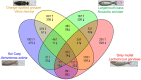
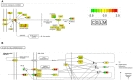
Transcriptome analysis is a powerful tool that enables a deep understanding of complicated physiological pathways, including immune responses. RNA sequencing (RNA-Seq)-based transcriptome analysis and various bioinformatics tools have also been used to study non-model animals, including aquaculture species for which reference genomes are not available. Rapid developments in these techniques have not only accelerated investigations into the process of pathogenic infection and defense strategies in fish, but also used to identify immunity-related genes in fish. These findings will contribute to fish immunotherapy for the prevention and treatment of bacterial infections through the design of more specific and effective immune stimulants, adjuvants, and vaccines. Until now, there has been little information regarding the universality and diversity of immune reactions against pathogenic infection in fish. Therefore, one of the aims of this paper is to introduce the RNA-Seq technique for examination of immune responses in pathogen-infected fish. This review also aims to highlight comparative studies of immune responses against bacteria, based on our previous findings in largemouth bass (Micropterus salmoides) against Nocardia seriolae, gray mullet (Mugil cephalus) against Lactococcus garvieae, orange-spotted grouper (Epinephelus coioides) against Vibrio harveyi, and koi carp (Cyprinus carpio) against Aeromonas sobria, using RNA-seq techniques. We demonstrated that only 39 differentially expressed genes (DEGs) were present in all species. However, the number of specific DEGs in each species was relatively higher than that of common DEGs; 493 DEGs in largemouth bass against N. seriolae, 819 DEGs in mullets against L. garvieae, 909 in groupers against V. harveyi, and 1471 in carps against A. sobria. The DEGs in different fish species were also representative of specific immune-related pathways. The results of this study will enhance our understanding of the immune responses of fish, and will aid in the development of effective vaccines, therapies, and disease-resistant strains.
Keywords: RNA-Seq; bacteria; fish disease; immune response; transcriptome.
Figures
Similar articles
-
Transcriptome analysis of immune response against Vibrio harveyi infection in orange-spotted grouper (Epinephelus coioides).Fish Shellfish Immunol. 2017 Nov;70:628-637. doi: 10.1016/j.fsi.2017.09.052. Epub 2017 Sep 20. Fish Shellfish Immunol. 2017. PMID: 28939531
-
Transcriptome analysis of grey mullet (Mugil cephalus) after challenge with Lactococcus garvieae.Fish Shellfish Immunol. 2016 Nov;58:593-603. doi: 10.1016/j.fsi.2016.10.006. Epub 2016 Oct 5. Fish Shellfish Immunol. 2016. PMID: 27720696
-
Immune-Related Functional Differential Gene Expression in Koi Carp (Cyprinus carpio) after Challenge with Aeromonas sobria.Int J Mol Sci. 2018 Jul 20;19(7):2107. doi: 10.3390/ijms19072107. Int J Mol Sci. 2018. PMID: 30036965 Free PMC article.
-
Transcriptome Analysis Based on RNA-Seq in Understanding Pathogenic Mechanisms of Diseases and the Immune System of Fish: A Comprehensive Review.Int J Mol Sci. 2018 Jan 15;19(1):245. doi: 10.3390/ijms19010245. Int J Mol Sci. 2018. PMID: 29342931 Free PMC article. Review.
-
miRNAs associated with immune response in teleost fish.Dev Comp Immunol. 2017 Oct;75:77-85. doi: 10.1016/j.dci.2017.02.023. Epub 2017 Feb 28. Dev Comp Immunol. 2017. PMID: 28254620 Review.
Cited by
-
PET-CT and RNA sequencing reveal novel targets for acupuncture-induced lowering of blood pressure in spontaneously hypertensive rats.Sci Rep. 2021 May 26;11(1):10973. doi: 10.1038/s41598-021-90467-1. Sci Rep. 2021. PMID: 34040073 Free PMC article.
-
Differential Expression Genes of the Head Kidney and Spleen in Streptococcus iniae-Infected East Asian Fourfinger Threadfin Fish (Eleutheronema tetradactylum).Int J Mol Sci. 2023 Feb 14;24(4):3832. doi: 10.3390/ijms24043832. Int J Mol Sci. 2023. PMID: 36835242 Free PMC article.
-
The Dynamic Immune Response of Yellow Catfish (Pelteobagrus fulvidraco) Infected With Edwardsiella ictaluri Presenting the Inflammation Process.Front Immunol. 2021 Feb 26;12:625928. doi: 10.3389/fimmu.2021.625928. eCollection 2021. Front Immunol. 2021. PMID: 33732247 Free PMC article.
-
Transcriptome Profiling of Pacu (Piaractus mesopotamicus) Challenged With Pathogenic Aeromonas hydrophila: Inference on Immune Gene Response.Front Genet. 2020 Jun 9;11:604. doi: 10.3389/fgene.2020.00604. eCollection 2020. Front Genet. 2020. PMID: 32582300 Free PMC article.
-
Transcriptomic Analysis of Fish Hosts Responses to Nervous Necrosis Virus.Pathogens. 2022 Feb 3;11(2):201. doi: 10.3390/pathogens11020201. Pathogens. 2022. PMID: 35215144 Free PMC article. Review.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Research Materials

