Laying the Foundation for Crassulacean Acid Metabolism (CAM) Biodesign: Expression of the C4 Metabolism Cycle Genes of CAM in Arabidopsis
- PMID: 30804970
- PMCID: PMC6378705
- DOI: 10.3389/fpls.2019.00101
Laying the Foundation for Crassulacean Acid Metabolism (CAM) Biodesign: Expression of the C4 Metabolism Cycle Genes of CAM in Arabidopsis
Abstract
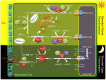
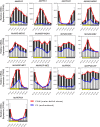
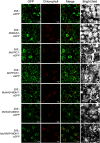
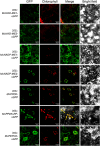
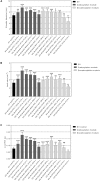
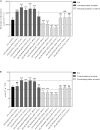
Crassulacean acid metabolism (CAM) is a specialized mode of photosynthesis that exploits a temporal CO2 pump with nocturnal CO2 uptake and concentration to reduce photorespiration, improve water-use efficiency (WUE), and optimize the adaptability of plants to hotter and drier climates. Introducing the CAM photosynthetic machinery into C3 (or C4) photosynthesis plants (CAM Biodesign) represents a potentially breakthrough strategy for improving WUE while maintaining high productivity. To optimize the success of CAM Biodesign approaches, the functional analysis of individual C4 metabolism cycle genes is necessary to identify the essential genes for robust CAM pathway introduction. Here, we isolated and analyzed the subcellular localizations of 13 enzymes and regulatory proteins of the C4 metabolism cycle of CAM from the common ice plant in stably transformed Arabidopsis thaliana. Six components of the carboxylation module were analyzed including beta-carbonic anhydrase (McBCA2), phosphoenolpyruvate carboxylase (McPEPC1), phosphoenolpyruvate carboxylase kinase (McPPCK1), NAD-dependent malate dehydrogenase (McNAD-MDH1, McNAD-MDH2), and NADP-dependent malate dehydrogenase (McNADP-MDH1). In addition, seven components of the decarboxylation module were analyzed including NAD-dependent malic enzyme (McNAD-ME1, McNAD-ME2), NADP-dependent malic enzyme (McNADP-ME1, NADP-ME2), pyruvate, orthophosphate dikinase (McPPDK), pyruvate, orthophosphate dikinase-regulatory protein (McPPDK-RP), and phosphoenolpyruvate carboxykinase (McPEPCK). Ectopic overexpression of most C4-metabolism cycle components resulted in increased rosette diameter, leaf area, and leaf fresh weight of A. thaliana except for McNADP-MDH1, McPPDK-RP, and McPEPCK. Overexpression of most carboxylation module components resulted in increased stomatal conductance and dawn/dusk titratable acidity (TA) as an indirect measure of organic acid (mainly malate) accumulation in A. thaliana. In contrast, overexpression of the decarboxylating malic enzymes reduced stomatal conductance and TA. This comprehensive study provides fundamental insights into the relative functional contributions of each of the individual components of the core C4-metabolism cycle of CAM and represents a critical first step in laying the foundation for CAM Biodesign.
Keywords: Arabidopsis thaliana; C4 metabolism; CAM biodesign; Mesembryanthemum crystallinum; crassulacean acid metabolism; ice plant; water-use efficiency.
Figures

References
-
- Araújo W. L., Nunes-Nesi A., Osorio S., Usadel B., Fuentes D., Nagy R., et al. (2011b). Antisense inhibition of the iron-sulphur subunit of succinate dehydrogenase enhances photosynthesis and growth in tomato via an organic acid-mediated effect on stomatal aperture. Plant Cell 23 600–627. 10.1105/tpc.110.081224 - DOI - PMC - PubMed
-
- Astley H. M., Parsley K., Aubry S., Chastain C. J., Burnell J. N., Webb M. E., et al. (2011). The pyruvate, orthophosphate dikinase regulatory proteins of Arabidopsis are both bifunctional and interact with the catalytic and nucleotide-binding domains of pyruvate, orthophosphate dikinase. Plant J. 68 1070–1080. 10.1111/j.1365-313X.2011.04759.x - DOI - PubMed
-
- Backhausen J. E., Emmerlich A., Holtgrefe S., Horton P., Nast G., Rogers J. J., et al. (1998). Transgenic potato plants with altered expression levels of chloroplast NADP-malate dehydrogenase: interactions between photosynthetic electron transport and malate metabolism in leaves and in isolated intact chloroplasts. Planta 207 105–114. 10.1007/s004250050461 - DOI
-
- Badia M. B., Arias C. L., Tronconi M. A., Maurino V. G., Andreo C. S., Drincovich M. F., et al. (2015). Enhanced cytosolic NADP-ME2 activity in A. thaliana affects plant development, stress tolerance and specific diurnal and nocturnal cellular processes. Plant Sci. 240 193–203. 10.1016/j.plantsci.2015.09.015 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

