Analysis of the genetic diversity and population structure of Salix psammophila based on phenotypic traits and simple sequence repeat markers
- PMID: 30805247
- PMCID: PMC6383557
- DOI: 10.7717/peerj.6419
Analysis of the genetic diversity and population structure of Salix psammophila based on phenotypic traits and simple sequence repeat markers
Abstract
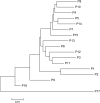
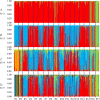
Salix psammophila (desert willow) is a shrub endemic to the Kubuqi Desert and the Mu Us Desert, China, that plays an important role in maintaining local ecosystems and can be used as a biomass feedstock for biofuels and bioenergy. However, the lack of information on phenotypic traits and molecular markers for this species limits the study of genetic diversity and population structure. In this study, nine phenotypic traits were analyzed to assess the morphological diversity and variation. The mean coefficient of variation of 17 populations ranged from 18.35% (branch angle (BA)) to 38.52% (leaf area (LA)). Unweighted pair-group method with arithmetic mean analysis of nine phenotypic traits of S. psammophila showed the same results, with the 17 populations clustering into five groups. We selected 491 genets of the 17 populations to analyze genetic diversity and population structure based on simple sequence repeat (SSR) markers. Analysis of molecular variance (AMOVA) revealed that most of the genetic variance (95%) was within populations, whereas only a small portion (5%) was among populations. Moreover, using the animal model with SSR-based relatedness estimated of S. psammophila, we found relatively moderate heritability values for phenotypic traits, suggesting that most of trait variation were caused by environmental or developmental variation. Principal coordinate and phylogenetic analyses based on SSR data revealed that populations P1, P2, P9, P16, and P17 were separated from the others. The results showed that the marginal populations located in the northeastern and southwestern had lower genetic diversity, which may be related to the direction of wind. These results provide a theoretical basis for germplasm management and genetic improvement of desert willow.
Keywords: Genetic diversity; Phenotypic traits; SSR; Salix psammophila; Structure genetics.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





Similar articles
-
De novo transcriptome assembly, development of EST-SSR markers and population genetic analyses for the desert biomass willow, Salix psammophila.Sci Rep. 2016 Dec 20;6:39591. doi: 10.1038/srep39591. Sci Rep. 2016. PMID: 27995985 Free PMC article.
-
Genetic diversity evaluation of Luculia yunnanensis, a vulnerable species endemic to Yunnan, Southwestern China based on morphological traits and EST-SSR markers.Front Plant Sci. 2024 Aug 14;15:1428364. doi: 10.3389/fpls.2024.1428364. eCollection 2024. Front Plant Sci. 2024. PMID: 39220013 Free PMC article.
-
Uncovering the genetic diversity of yams (Dioscorea spp.) in China by combining phenotypic trait and molecular marker analyses.Ecol Evol. 2021 Jul 16;11(15):9970-9986. doi: 10.1002/ece3.7727. eCollection 2021 Aug. Ecol Evol. 2021. PMID: 34367553 Free PMC article.
-
Genetic Diversity Study on Geographical Populations of the Multipurpose Species Elsholtzia stauntonii Using Transferable Microsatellite Markers.Front Plant Sci. 2022 May 12;13:903674. doi: 10.3389/fpls.2022.903674. eCollection 2022. Front Plant Sci. 2022. PMID: 35646027 Free PMC article.
-
Phenotype- and SSR-Based Estimates of Genetic Variation between and within Two Important Elymus Species in Western and Northern China.Genes (Basel). 2018 Mar 7;9(3):147. doi: 10.3390/genes9030147. Genes (Basel). 2018. PMID: 29518961 Free PMC article.
Cited by
-
Phenotypic diversity and provenance variation of Cupressus funebris: a case study in the Sichuan Basin, China.PeerJ. 2024 Nov 29;12:e18494. doi: 10.7717/peerj.18494. eCollection 2024. PeerJ. 2024. PMID: 39624132 Free PMC article.
-
Insights into Molecular Mechanism of Secondary Xylem Rapid Growth in Salix psammophila.Plants (Basel). 2025 Feb 5;14(3):459. doi: 10.3390/plants14030459. Plants (Basel). 2025. PMID: 39943021 Free PMC article.
-
Genetic Structure and Conservation Management of Endemic Salix kusanoi in Fragmented Habitats of Taiwan.Plants (Basel). 2025 Apr 1;14(7):1080. doi: 10.3390/plants14071080. Plants (Basel). 2025. PMID: 40219147 Free PMC article.
-
Assessment of the Genetic Relationship and Population Structure in Oil-Tea Camellia Species Using Simple Sequence Repeat (SSR) Markers.Genes (Basel). 2022 Nov 19;13(11):2162. doi: 10.3390/genes13112162. Genes (Basel). 2022. PMID: 36421835 Free PMC article.
-
Genome-wide identification and molecular evolution of Dof gene family in Camellia oleifera.BMC Genomics. 2024 Jul 18;25(1):702. doi: 10.1186/s12864-024-10622-6. BMC Genomics. 2024. PMID: 39026173 Free PMC article.
References
-
- Bao Y, Zhang G. Study of adsorption characteristics of methylene blue onto activated carbon made by Salix Psammophila. Energy Procedia. 2012;16:1141–1146. doi: 10.1016/j.egypro.2012.01.182. - DOI
-
- Barker JHA, Pahlich A, Trybush S, Edwards KJ, Karp A. Microsatellite markers for diverse Salix species. Molecular Ecology Resources. 2010;3(1):4–6. doi: 10.1046/j.1471-8286.2003.00332.x. - DOI
-
- Bates D, Maechler M, Bolker B, Walker S. lme4: linear mixed effects models using eigen and S4. R package version 1.0-6http://CRAN.R-project.org/package=lme4 2014
-
- Berlin S, Trybush SO, Fogelqvist J, Gyllenstrand N, Hallingbäck HR, Åhman I, Nordh NE, Shield I, Powers SJ, Weih M, Lagercrantz U, Rönnberg-Wästljung AC, Karp A, Hanley SJ. Genetic diversity, population structure and phenotypic variation in European Salix viminalis L. (Salicaceae) Tree Genetics & Genomes. 2014;10(6):1595–1610. doi: 10.1007/s11295-014-0782-5. - DOI
-
- Bozzi JA, Liepelt S, Ohneiser S, Gallo LA, Marchelli P, Leyer I, Ziegenhagen B, Mengel C. Characterization of 23 polymorphic SSR markers in Salix humboldtiana (Salicaceae) using next-generation sequencing and cross-amplification from related species. Applications in Plant Science. 2015;3(4):1400120. doi: 10.3732/apps.1400120. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

