DeepSSM: A Deep Learning Framework for Statistical Shape Modeling from Raw Images
- PMID: 30805572
- PMCID: PMC6385885
- DOI: 10.1007/978-3-030-04747-4_23
DeepSSM: A Deep Learning Framework for Statistical Shape Modeling from Raw Images
Abstract
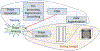
Statistical shape modeling is an important tool to characterize variation in anatomical morphology. Typical shapes of interest are measured using 3D imaging and a subsequent pipeline of registration, segmentation, and some extraction of shape features or projections onto some lower-dimensional shape space, which facilitates subsequent statistical analysis. Many methods for constructing compact shape representations have been proposed, but are often impractical due to the sequence of image preprocessing operations, which involve significant parameter tuning, manual delineation, and/or quality control by the users. We propose DeepSSM: a deep learning approach to extract a low-dimensional shape representation directly from 3D images, requiring virtually no parameter tuning or user assistance. DeepSSM uses a convolutional neural network (CNN) that simultaneously localizes the biological structure of interest, establishes correspondences, and projects these points onto a low-dimensional shape representation in the form of PCA loadings within a point distribution model. To overcome the challenge of the limited availability of training images with dense correspondences, we present a novel data augmentation procedure that uses existing correspondences on a relatively small set of processed images with shape statistics to create plausible training samples with known shape parameters. In this way, we leverage the limited CT/MRI scans (40-50) into thousands of images needed to train a deep neural net. After the training, the CNN automatically produces accurate low-dimensional shape representations for unseen images. We validate DeepSSM for three different applications pertaining to modeling pediatric cranial CT for characterization of metopic craniosynostosis, femur CT scans identifying morphologic deformities of the hip due to femoroacetabular impingement, and left atrium MRI scans for atrial fibrillation recurrence prediction.
Figures
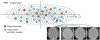
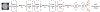





Similar articles
-
DeepSSM: A blueprint for image-to-shape deep learning models.Med Image Anal. 2024 Jan;91:103034. doi: 10.1016/j.media.2023.103034. Epub 2023 Nov 17. Med Image Anal. 2024. PMID: 37984127 Free PMC article.
-
Progressive DeepSSM: Training Methodology for Image-To-Shape Deep Models.Shape Med Imaging (2023). 2023 Oct;14350:157-172. doi: 10.1007/978-3-031-46914-5_13. Epub 2023 Oct 31. Shape Med Imaging (2023). 2023. PMID: 38745942 Free PMC article.
-
Uncertain-DeepSSM: From Images to Probabilistic Shape Models.Shape Med Imaging (2020). 2020 Oct;12474:57-72. doi: 10.1007/978-3-030-61056-2_5. Epub 2020 Oct 3. Shape Med Imaging (2020). 2020. PMID: 33817703 Free PMC article.
-
Image2SSM: Reimagining Statistical Shape Models from Images with Radial Basis Functions.Med Image Comput Comput Assist Interv. 2023 Oct;14220:508-517. doi: 10.1007/978-3-031-43907-0_49. Epub 2023 Oct 1. Med Image Comput Comput Assist Interv. 2023. PMID: 39534229 Free PMC article.
-
Automatic Segmentation of Multiple Organs on 3D CT Images by Using Deep Learning Approaches.Adv Exp Med Biol. 2020;1213:135-147. doi: 10.1007/978-3-030-33128-3_9. Adv Exp Med Biol. 2020. PMID: 32030668 Review.
Cited by
-
How machine learning is impacting research in atrial fibrillation: implications for risk prediction and future management.Cardiovasc Res. 2021 Jun 16;117(7):1700-1717. doi: 10.1093/cvr/cvab169. Cardiovasc Res. 2021. PMID: 33982064 Free PMC article. Review.
-
Generating Synthetic Labeled Data From Existing Anatomical Models: An Example With Echocardiography Segmentation.IEEE Trans Med Imaging. 2021 Oct;40(10):2783-2794. doi: 10.1109/TMI.2021.3051806. Epub 2021 Sep 30. IEEE Trans Med Imaging. 2021. PMID: 33444134 Free PMC article.
-
MASSM: An End-to-End Deep Learning Framework for Multi-Anatomy Statistical Shape Modeling Directly From Images.Shape Med Imaging (2024). 2025;15275:149-163. doi: 10.1007/978-3-031-75291-9_12. Epub 2024 Oct 26. Shape Med Imaging (2024). 2025. PMID: 39649703
-
Weakly Supervised Bayesian Shape Modeling from Unsegmented Medical Images.Shape Med Imaging (2024). 2025;15275:1-17. Epub 2024 Oct 26. Shape Med Imaging (2024). 2025. PMID: 39605948
-
Automating Linear and Angular Measurements for the Hip and Knee After Computed Tomography: Validation of a Three-Stage Deep Learning and Computer Vision-Based Pipeline for Pathoanatomic Assessment.Arthroplast Today. 2024 May 11;27:101394. doi: 10.1016/j.artd.2024.101394. eCollection 2024 Jun. Arthroplast Today. 2024. PMID: 39071819 Free PMC article.
References
-
- Abadi M, Barham P, Chen J, Chen Z, Davis A, Dean J, Devin M, Ghemawat S, Irving G, Isard M, et al.: Tensorflow: A system for large-scale machine learning. In: OSDI. vol. 16, pp. 265–283 (2016)
-
- Atkins PR, Elhabian SY, Agrawal P, Harris MD, Whitaker RT, Weiss JA, Peters CL, Anderson AE: Quantitative comparison of cortical bone thickness using correspondence-based shape modeling in patients with cam femoroacetabular impingement. Journal of Orthopaedic Research 35(8), 1743–1753 (2017) - PMC - PubMed
-
- Badrinarayanan V, Handa A, Cipolla R: Segnet: A deep convolutional encoder-decoder architecture for robust semantic pixel-wise labelling. arXiv preprint arXiv:1505.07293 (2015)
-
- Beg MF, Miller MI, Trouvé A, Younes L: Computing large deformation metric mappings via geodesic flows of diffeomorphisms. International journal of computer vision 61(2), 139–157 (2005)
-
- Bieging ET, Morris A, Wilson BD, McGann CJ, Marrouche NF, Cates J: Left atrial shape predicts recurrence after atrial fibrillation catheter ablation. Journal of cardiovascular electrophysiology (2018) - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
