Quantitative proteomics identifies proteins that resist translational repression and become dysregulated in ALS-FUS
- PMID: 30806671
- PMCID: PMC6586143
- DOI: 10.1093/hmg/ddz048
Quantitative proteomics identifies proteins that resist translational repression and become dysregulated in ALS-FUS
Abstract
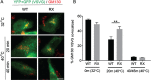
Aberrant translational repression is a feature of multiple neurodegenerative diseases. The association between disease-linked proteins and stress granules further implicates impaired stress responses in neurodegeneration. However, our knowledge of the proteins that evade translational repression is incomplete. It is also unclear whether disease-linked proteins influence the proteome under conditions of translational repression. To address these questions, a quantitative proteomics approach was used to identify proteins that evade stress-induced translational repression in arsenite-treated cells expressing either wild-type or amyotrophic lateral sclerosis (ALS)-linked mutant FUS. This study revealed hundreds of proteins that are actively synthesized during stress-induced translational repression, irrespective of FUS genotype. In addition to proteins involved in RNA- and protein-processing, proteins associated with neurodegenerative diseases such as ALS were also actively synthesized during stress. Protein synthesis under stress was largely unperturbed by mutant FUS, although several proteins were found to be differentially expressed between mutant and control cells. One protein in particular, COPBI, was downregulated in mutant FUS-expressing cells under stress. COPBI is the beta subunit of the coat protein I (COPI), which is involved in Golgi to endoplasmic reticulum (ER) retrograde transport. Further investigation revealed reduced levels of other COPI subunit proteins and defects in COPBI-relatedprocesses in cells expressing mutant FUS. Even in the absence of stress, COPBI localization was altered in primary and human stem cell-derived neurons expressing ALS-linked FUS variants. Our results suggest that Golgi to ER retrograde transport may be important under conditions of stress and is perturbed upon the expression of disease-linked proteins such as FUS.
© The Author(s) 2019. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures

Similar articles
-
Ataxin-2 interacts with FUS and intermediate-length polyglutamine expansions enhance FUS-related pathology in amyotrophic lateral sclerosis.Hum Mol Genet. 2013 Feb 15;22(4):717-28. doi: 10.1093/hmg/dds479. Epub 2012 Nov 19. Hum Mol Genet. 2013. PMID: 23172909
-
Rab1-dependent ER-Golgi transport dysfunction is a common pathogenic mechanism in SOD1, TDP-43 and FUS-associated ALS.Acta Neuropathol. 2015 Nov;130(5):679-97. doi: 10.1007/s00401-015-1468-2. Epub 2015 Aug 23. Acta Neuropathol. 2015. PMID: 26298469
-
ALS mutations of FUS suppress protein translation and disrupt the regulation of nonsense-mediated decay.Proc Natl Acad Sci U S A. 2018 Dec 18;115(51):E11904-E11913. doi: 10.1073/pnas.1810413115. Epub 2018 Nov 19. Proc Natl Acad Sci U S A. 2018. PMID: 30455313 Free PMC article.
-
Stress granules in neurodegeneration--lessons learnt from TAR DNA binding protein of 43 kDa and fused in sarcoma.FEBS J. 2013 Sep;280(18):4348-70. doi: 10.1111/febs.12287. Epub 2013 May 9. FEBS J. 2013. PMID: 23587065 Review.
-
Stress Granules and ALS: A Case of Causation or Correlation?Adv Neurobiol. 2018;20:173-212. doi: 10.1007/978-3-319-89689-2_7. Adv Neurobiol. 2018. PMID: 29916020 Review.
Cited by
-
Implications of mRNA translation dysregulation for neurological disorders.Semin Cell Dev Biol. 2021 Jun;114:11-19. doi: 10.1016/j.semcdb.2020.09.005. Epub 2020 Oct 21. Semin Cell Dev Biol. 2021. PMID: 34024497 Free PMC article. Review.
-
Advanced imaging techniques for studying protein phase separation in living cells and at single-molecule level.Curr Opin Chem Biol. 2023 Oct;76:102371. doi: 10.1016/j.cbpa.2023.102371. Epub 2023 Jul 29. Curr Opin Chem Biol. 2023. PMID: 37523989 Free PMC article. Review.
-
G3BP1-linked mRNA partitioning supports selective protein synthesis in response to oxidative stress.Nucleic Acids Res. 2020 Jul 9;48(12):6855-6873. doi: 10.1093/nar/gkaa376. Nucleic Acids Res. 2020. PMID: 32406909 Free PMC article.
-
FUS regulates autophagy by mediating the transcription of genes critical to the autophagosome formation.J Neurochem. 2021 May;157(3):752-763. doi: 10.1111/jnc.15281. Epub 2021 Jan 18. J Neurochem. 2021. PMID: 33354770 Free PMC article.
-
Interactions between ALS-linked FUS and nucleoporins are associated with defects in the nucleocytoplasmic transport pathway.Nat Neurosci. 2021 Aug;24(8):1077-1088. doi: 10.1038/s41593-021-00859-9. Epub 2021 May 31. Nat Neurosci. 2021. PMID: 34059832 Free PMC article.
References
-
- Holcik M. and Sonenberg N. (2005) Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol., 6, 318–327. - PubMed
-
- Souquere S., Mollet S., Kress M., Dautry F., Pierron G. and Weil D. (2009) Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci., 122, 3619–3626. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

