Proteomic Analysis Reveals Novel Mechanisms by Which Polychlorinated Biphenyls Compromise the Liver Promoting Diet-Induced Steatohepatitis
- PMID: 30807179
- PMCID: PMC7059562
- DOI: 10.1021/acs.jproteome.8b00886
Proteomic Analysis Reveals Novel Mechanisms by Which Polychlorinated Biphenyls Compromise the Liver Promoting Diet-Induced Steatohepatitis
Abstract
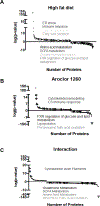
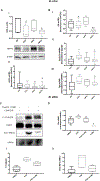
Environmental pollution contributes to fatty liver disease pathogenesis. Polychlorinated biphenyl (PCB) exposures have been associated with liver enzyme elevation and suspected steatohepatitis in cohort studies. Male mice treated with the commercial PCB mixture, Aroclor 1260 (20 mg/kg), and fed high fat diet (HFD) for 12 weeks developed steatohepatitis. Receptor-based modes of action including inhibition of the epidermal growth factor (EGF) receptor were previously proposed, but other mechanisms likely exist. Objectives were to identify and validate the pathways, transcription factors, and mechanisms responsible for the steatohepatitis associated with PCB and HFD coexposures. Comparative proteomics analysis was performed in archived mouse liver samples from the aforementioned chronic exposure study. Pathway and transcription factor analysis (TFA) was performed, and selected results were validated. Liver proteomics detected 1103 unique proteins. Aroclor 1260 upregulated 154 and downregulated 93 of these. Aroclor 1260 + HFD coexposures affected 55 pathways including glutathione metabolism, intermediary metabolism, and cytoskeletal remodeling. TFA of Aroclor 1260 treatment demonstrated alterations in the function of 42 transcription factors including downregulation of NRF2 and key nuclear receptors previously demonstrated to protect against steatohepatitis (e.g., HNF4α, FXR, PPARα/δ/γ, etc.). Validation studies demonstrated that Aroclor 1260 significantly reduced HNF4α protein levels, while Aroclor 1260 + HFD reduced expression of the HNF4α target gene, albumin, in vivo. Aroclor 1260 attenuated EGF-dependent HNF4α phosphorylation and target gene activation in vitro. Aroclor 1260 reduced levels of NRF2, its target genes, and glutathione in vivo. Aroclor 1260 attenuated EGF-dependent NRF2 upregulation, in vitro. Aroclor 1260 indirectly activated hepatic stellate cells in vitro via induction of hepatocyte-derived TGFβ. PCB exposures adversely impacted transcription factors regulating liver protection, function, and fibrosis. PCBs, thus, compromised the liver by reducing its protective responses against nutritional stress to promote diet-induced steatohepatitis. The identified mechanisms by which environmental pollutants influence fatty liver disease pathogenesis require confirmation in humans.
Keywords: Aroclor 1260; EGFR; HNF4α; TASH; liver; metabolism; polychlorinated biphenyl; steatohepatitis.
Figures



References
-
- Younossi ZM; Koenig AB; Abdelatif D; Fazel Y; Henry L; Wymer M, Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016, 64, (1), 73–84. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

