A review of 10 years of human microbiome research activities at the US National Institutes of Health, Fiscal Years 2007-2016
- PMID: 30808411
- PMCID: PMC6391833
- DOI: 10.1186/s40168-019-0620-y
A review of 10 years of human microbiome research activities at the US National Institutes of Health, Fiscal Years 2007-2016
Abstract
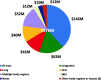
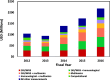
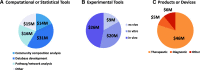
The National Institutes of Health (NIH) is the primary federal government agency for biomedical research in the USA. NIH provides extensive support for human microbiome research with 21 of 27 NIH Institutes and Centers (ICs) currently funding this area through their extramural research programs. This analysis of the NIH extramural portfolio in human microbiome research briefly reviews the early history of this field at NIH, summarizes the program objectives and the resources developed in the recently completed 10-year (fiscal years 2007-2016) $215 M Human Microbiome Project (HMP) program, evaluates the scope and range of the $728 M NIH investment in extramural human microbiome research activities outside of the HMP over fiscal years 2012-2016, and highlights some specific areas of research which emerged from this investment. This analysis closes with a few comments on the technical needs and knowledge gaps which remain for this field to be able to advance over the next decade and for the outcomes of this research to be able to progress to microbiome-based interventions for treating disease and supporting health.
Keywords: Animal models; Fast Track Action Committee on Mapping the Microbiome (FTAC-MM); Human Microbiome Project (HMP); Human disease; Metagenomics; Microbiome; Microbiota; NIH funding; National Microbiome Initiative (NMI).
Conflict of interest statement
Competing interests
The authors declare that they have no competing interests. The views expressed in this manuscript are those of the authors and not of the NIH or HHS.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
-
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–3.10.1038/4441022a - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

