Efficient Implementation of Penalized Regression for Genetic Risk Prediction
- PMID: 30808621
- PMCID: PMC6499521
- DOI: 10.1534/genetics.119.302019
Efficient Implementation of Penalized Regression for Genetic Risk Prediction
Abstract
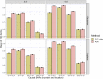
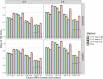
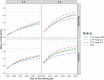
Polygenic Risk Scores (PRS) combine genotype information across many single-nucleotide polymorphisms (SNPs) to give a score reflecting the genetic risk of developing a disease. PRS might have a major impact on public health, possibly allowing for screening campaigns to identify high-genetic risk individuals for a given disease. The "Clumping+Thresholding" (C+T) approach is the most common method to derive PRS. C+T uses only univariate genome-wide association studies (GWAS) summary statistics, which makes it fast and easy to use. However, previous work showed that jointly estimating SNP effects for computing PRS has the potential to significantly improve the predictive performance of PRS as compared to C+T. In this paper, we present an efficient method for the joint estimation of SNP effects using individual-level data, allowing for practical application of penalized logistic regression (PLR) on modern datasets including hundreds of thousands of individuals. Moreover, our implementation of PLR directly includes automatic choices for hyper-parameters. We also provide an implementation of penalized linear regression for quantitative traits. We compare the performance of PLR, C+T and a derivation of random forests using both real and simulated data. Overall, we find that PLR achieves equal or higher predictive performance than C+T in most scenarios considered, while being scalable to biobank data. In particular, we find that improvement in predictive performance is more pronounced when there are few effects located in nearby genomic regions with correlated SNPs; for instance, in simulations, AUC values increase from 83% with the best prediction of C+T to 92.5% with PLR. We confirm these results in a data analysis of a case-control study for celiac disease where PLR and the standard C+T method achieve AUC values of 89% and of 82.5%. Applying penalized linear regression to 350,000 individuals of the UK Biobank, we predict height with a larger correlation than with the best prediction of C+T (∼65% instead of ∼55%), further demonstrating its scalability and strong predictive power, even for highly polygenic traits. Moreover, using 150,000 individuals of the UK Biobank, we are able to predict breast cancer better than C+T, fitting PLR in a few minutes only. In conclusion, this paper demonstrates the feasibility and relevance of using penalized regression for PRS computation when large individual-level datasets are available, thanks to the efficient implementation available in our R package bigstatsr.
Keywords: GenPred; LASSO; SNP; genomic prediction; polygenic risk scores; shared data resources.
Copyright © 2019 Privé et al.
Figures
References
-
- Breiman L., 2001. Random forests. Mach. Learn. 45: 5–32. 10.1023/A:1010933404324 - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

