A mutually induced conformational fit underlies Ca2+-directed interactions between calmodulin and the proximal C terminus of KCNQ4 K+ channels
- PMID: 30808708
- PMCID: PMC6463706
- DOI: 10.1074/jbc.RA118.006857
A mutually induced conformational fit underlies Ca2+-directed interactions between calmodulin and the proximal C terminus of KCNQ4 K+ channels
Abstract
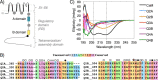
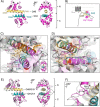
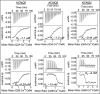
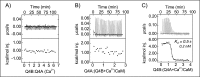
Calmodulin (CaM) conveys intracellular Ca2+ signals to KCNQ (Kv7, "M-type") K+ channels and many other ion channels. Whether this "calmodulation" involves a dramatic structural rearrangement or only slight perturbations of the CaM/KCNQ complex is as yet unclear. A consensus structural model of conformational shifts occurring between low nanomolar and physiologically high intracellular [Ca2+] is still under debate. Here, we used various techniques of biophysical chemical analyses to investigate the interactions between CaM and synthetic peptides corresponding to the A and B domains of the KCNQ4 subtype. We found that in the absence of CaM, the peptides are disordered, whereas Ca2+/CaM imposed helical structure on both KCNQ A and B domains. Isothermal titration calorimetry revealed that Ca2+/CaM has higher affinity for the B domain than for the A domain of KCNQ2-4 and much higher affinity for the B domain when prebound with the A domain. X-ray crystallography confirmed that these discrete peptides spontaneously form a complex with Ca2+/CaM, similar to previous reports of CaM binding KCNQ-AB domains that are linked together. Microscale thermophoresis and heteronuclear single-quantum coherence NMR spectroscopy indicated the C-lobe of Ca2+-free CaM to interact with the KCNQ4 B domain (Kd ∼10-20 μm), with increasing Ca2+ molar ratios shifting the CaM-B domain interactions via only the CaM C-lobe to also include the N-lobe. Our findings suggest that in response to increased Ca2+, CaM undergoes lobe switching that imposes a dramatic mutually induced conformational fit to both the proximal C terminus of KCNQ4 channels and CaM, likely underlying Ca2+-dependent regulation of KCNQ gating.
Keywords: M current; X-ray crystallography; biophysical chemical analysis; calcium signaling; calcium-binding protein; calmodulin (CaM); conformational change; isothermal titration calorimetry (ITC); microscale thermophoresis (MST); nuclear magnetic resonance (NMR); potassium channel; potassium voltage-gated channel subfamily Q member (KCNQ).
© 2019 Archer et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures






References
-
- Linse S., Helmersson A., and Forsén S. (1991) Calcium binding to calmodulin and its globular domains. J. Biol. Chem. 266, 8050–8054 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
- F31 NS090887/NS/NINDS NIH HHS/United States
- R01 NS043394/NS/NINDS NIH HHS/United States
- S10 OD021527/OD/NIH HHS/United States
- T32 HL007446/HL/NHLBI NIH HHS/United States
- R01 NS094461/NS/NINDS NIH HHS/United States
- R01 NS065138/NS/NINDS NIH HHS/United States
- P30 GM124165/GM/NIGMS NIH HHS/United States
- R01 AI104476/AI/NIAID NIH HHS/United States
- K12 GM111726/GM/NIGMS NIH HHS/United States
- R56 NS065138/NS/NINDS NIH HHS/United States
- P30 CA054174/CA/NCI NIH HHS/United States
- UL1 TR001120/TR/NCATS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Miscellaneous

