Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin
- PMID: 30808726
- PMCID: PMC6396422
- DOI: 10.1101/gr.242719.118
Orientation-aware plasma cell-free DNA fragmentation analysis in open chromatin regions informs tissue of origin
Abstract
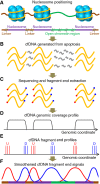
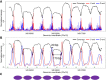
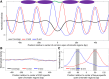
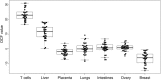
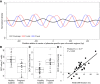
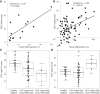
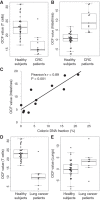
Cell-free DNA (cfDNA) in human plasma is a class of biomarkers with many current and potential future diagnostic applications. Recent studies have shown that cfDNA molecules are not randomly fragmented and possess information related to their tissues of origin. Pathologies causing death of cells from particular tissues result in perturbations in the relative distribution of DNA from the affected tissues. Such tissue-of-origin analysis is particularly useful in the development of liquid biopsies for cancer. It is therefore of value to accurately determine the relative contributions of the tissues to the plasma DNA pool in a simultaneous manner. In this work, we report that in open chromatin regions, cfDNA molecules show characteristic fragmentation patterns reflected by sequencing coverage imbalance and differentially phased fragment end signals. The latter refers to differences in the read densities of sequences corresponding to the orientation of the upstream and downstream ends of cfDNA molecules in relation to the reference genome. Such cfDNA fragmentation patterns preferentially occur in tissue-specific open chromatin regions where the corresponding tissues contributed DNA into the plasma. Quantitative analyses of such signals allow measurement of the relative contributions of various tissues toward the plasma DNA pool. These findings were validated by plasma DNA sequencing data obtained from pregnant women, organ transplantation recipients, and cancer patients. Orientation-aware plasma DNA fragmentation analysis therefore has potential diagnostic applications in noninvasive prenatal testing, organ transplantation monitoring, and cancer liquid biopsy.
© 2019 Sun et al.; Published by Cold Spring Harbor Laboratory Press.
Figures
References
-
- Chan KCA, Jiang P, Chan CW, Sun K, Wong J, Hui EP, Chan SL, Chan WC, Hui DS, Ng SS, et al. 2013a. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci 110: 18761–18768. 10.1073/pnas.1313995110 - DOI - PMC - PubMed
-
- Chan KCA, Jiang P, Zheng YW, Liao GJ, Sun H, Wong J, Siu SS, Chan WC, Chan SL, Chan AT, et al. 2013b. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 59: 211–224. 10.1373/clinchem.2012.196014 - DOI - PubMed
-
- Chan KCA, Jiang P, Sun K, Cheng YK, Tong YK, Cheng SH, Wong AI, Hudecova I, Leung TY, Chiu RW, et al. 2016. Second generation noninvasive fetal genome analysis reveals de novo mutations, single-base parental inheritance, and preferred DNA ends. Proc Natl Acad Sci 113: E8159–E8168. 10.1073/pnas.1615800113 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
