Simultaneous Bayesian inference of phylogeny and molecular coevolution
- PMID: 30808804
- PMCID: PMC6421416
- DOI: 10.1073/pnas.1813836116
Simultaneous Bayesian inference of phylogeny and molecular coevolution
Abstract
Patterns of molecular coevolution can reveal structural and functional constraints within or among organic molecules. These patterns are better understood when considering the underlying evolutionary process, which enables us to disentangle the signal of the dependent evolution of sites (coevolution) from the effects of shared ancestry of genes. Conversely, disregarding the dependent evolution of sites when studying the history of genes negatively impacts the accuracy of the inferred phylogenetic trees. Although molecular coevolution and phylogenetic history are interdependent, analyses of the two processes are conducted separately, a choice dictated by computational convenience, but at the expense of accuracy. We present a Bayesian method and associated software to infer how many and which sites of an alignment evolve according to an independent or a pairwise dependent evolutionary process, and to simultaneously estimate the phylogenetic relationships among sequences. We validate our method on synthetic datasets and challenge our predictions of coevolution on the 16S rRNA molecule by comparing them with its known molecular structure. Finally, we assess the accuracy of phylogenetic trees inferred under the assumption of independence among sites using synthetic datasets, the 16S rRNA molecule and 10 additional alignments of protein-coding genes of eukaryotes. Our results demonstrate that inferring phylogenetic trees while accounting for dependent site evolution significantly impacts the estimates of the phylogeny and the evolutionary process.
Keywords: Bayesian inference; molecular coevolution; phylogeny; tree of life.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
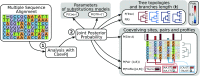

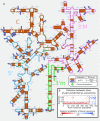


References
-
- de Juan D, Pazos F, Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet. 2013;14:249–261. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

