Inbreeding load and inbreeding depression estimated from lifetime reproductive success in a small, dispersal-limited population
- PMID: 30809076
- PMCID: PMC6781135
- DOI: 10.1038/s41437-019-0197-z
Inbreeding load and inbreeding depression estimated from lifetime reproductive success in a small, dispersal-limited population
Abstract
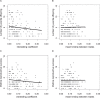
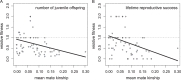
The fitness consequences of inbreeding and the individual behaviors that prevent its detrimental effects can be challenging to document in wild populations. Here, we use field and molecular data from a 17-year study of banner-tailed kangaroo rats (Dipodomys spectabilis) to quantify the relationship between inbreeding, mate kinship, and lifetime reproductive success. Using a pedigree that was reconstructed using genetic and field data within a Bayesian framework (median probability of parental assignment = 0.92, mean pedigree depth = 6 generations), we estimated both inbreeding coefficients and kinship between individuals that produced offspring (mean inbreeding coefficient = 0.07, mean mate kinship = 0.08). We also used the pedigree, in combination with census data, to generate a series of fitness estimates, ranging from survival to reproductive maturity to lifetime reproductive success. We found that the population's inbreeding load was low to moderate (0.98-4.66 haploid lethal equivalents) and increased with the time frame over which fitness was estimated (lowest for survival to maturity, highest for adult-to-adult reproductive success). Fitness decreased with increasing inbreeding coefficients. For example, lifetime reproductive success was reduced by 24% for individuals with inbreeding coefficients greater than twice the population mean. Within full sibling pairs, the sibling with less-related mates produced an average of 30% more offspring over its lifetime. These data further illustrate that inbreeding can have a negative effect on lifetime reproductive success.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
-
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2014;67:1–48.
-
- Best TL. Mound development by a pioneer population of the banner-tailed kangaroo rat, Dipodomys spectabilis baileyi Goldman, in Eastern New Mexico. Am Midl Nat. 1972;87:201–206. doi: 10.2307/2423893. - DOI
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

