Transcriptional Shifts Highlight the Role of Nutrients in Harmful Brown Tide Dynamics
- PMID: 30809203
- PMCID: PMC6379262
- DOI: 10.3389/fmicb.2019.00136
Transcriptional Shifts Highlight the Role of Nutrients in Harmful Brown Tide Dynamics
Abstract
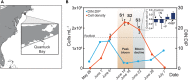
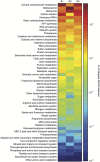
Harmful algal blooms (HABs) threaten ecosystems and human health worldwide. Controlling nitrogen inputs to coastal waters is a common HAB management strategy, as nutrient concentrations often suggest coastal blooms are nitrogen-limited. However, defining best nutrient management practices is a long-standing challenge: in part, because of difficulties in directly tracking the nutritional physiology of harmful species in mixed communities. Using metatranscriptome sequencing and incubation experiments, we addressed this challenge by assaying the in situ physiological ecology of the ecosystem destructive alga, Aureococcus anophagefferens. Here we show that gene markers of phosphorus deficiency were expressed in situ, and modulated by the enrichment of phosphorus, which was consistent with the observed growth rate responses. These data demonstrate the importance of phosphorus in controlling brown-tide dynamics, suggesting that phosphorus, in addition to nitrogen, should be evaluated in the management and mitigation of these blooms. Given that nutrient concentrations alone were suggestive of a nitrogen-limited ecosystem, this study underscores the value of directly assaying harmful algae in situ for the development of management strategies.
Keywords: Aureococcus anophagefferens; brown tide; harmful algal bloom; metatranscriptomics; nutrient physiology.
Figures


References
-
- Anderson D. M., Glibert P. M., Burkholder J. M. (2002). Harmful algal blooms and eutrophication: nutrient sources, composition, and consequences. Estuaries 25 704–726. 10.1007/BF02804901 - DOI
LinkOut - more resources
Full Text Sources

