Characterization of Clinically Relevant Strains of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Occurring in Environmental Sources in a Rural Area of China by Using Whole-Genome Sequencing
- PMID: 30809212
- PMCID: PMC6379450
- DOI: 10.3389/fmicb.2019.00211
Characterization of Clinically Relevant Strains of Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae Occurring in Environmental Sources in a Rural Area of China by Using Whole-Genome Sequencing
Abstract
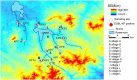
Klebsiella pneumoniae is a gram-negative, opportunistic pathogen, and a common cause of healthcare-associated infections such as pneumonia, septicemia, and urinary tract infection. The purpose of this study was to survey the occurrence of and characterize K. pneumoniae in different environmental sources in a rural area of Shandong province, China. Two hundred and thirty-one samples from different environmental sources in 12 villages were screened for extended-spectrum β-lactamase-(ESBL)-producing K. pneumoniae, and 14 (6%) samples were positive. All isolates were multidrug-resistant and a few of them belonged to clinically relevant strains which are known to cause hospital outbreaks worldwide. Serotypes, virulence genes, serum survival, and phagocytosis survival were analyzed and the results showed the presence of virulence factors associated with highly virulent clones and a high degree of phagocytosis survivability, indicating the potential virulence of these isolates. These results emphasize the need for further studies designed to elucidate the role of the environment in transmission and dissemination of ESBL-producing K. pneumoniae and the potential risk posed to human and environmental health.
Keywords: Klebsiella pneumoniae; environment; extended-spectrum β-lactamase; feces; multilocus sequence typing; pulsed-field gel electrophoresis; water; whole-genome sequencing.
Figures


Similar articles
-
Molecular characteristics of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae causing intra-abdominal infections from 9 tertiary hospitals in China.Diagn Microbiol Infect Dis. 2017 Jan;87(1):45-48. doi: 10.1016/j.diagmicrobio.2016.10.007. Epub 2016 Oct 7. Diagn Microbiol Infect Dis. 2017. PMID: 27773544
-
Phenotypic and Molecular Characterization of Antimicrobial Resistance in Klebsiella spp. Isolates from Companion Animals in Japan: Clonal Dissemination of Multidrug-Resistant Extended-Spectrum β-Lactamase-Producing Klebsiella pneumoniae.Front Microbiol. 2016 Jun 29;7:1021. doi: 10.3389/fmicb.2016.01021. eCollection 2016. Front Microbiol. 2016. PMID: 27446056 Free PMC article.
-
Whole-Genome Sequencing of Human Clinical Klebsiella pneumoniae Isolates Reveals Misidentification and Misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae.mSphere. 2017 Aug 2;2(4):e00290-17. doi: 10.1128/mSphereDirect.00290-17. eCollection 2017 Jul-Aug. mSphere. 2017. PMID: 28776045 Free PMC article.
-
Molecular characterisation and epidemiology of extended-spectrum β-lactamase-producing Klebsiella pneumoniae isolates from immunocompromised patients in Tunisia.J Glob Antimicrob Resist. 2018 Jun;13:154-160. doi: 10.1016/j.jgar.2017.12.014. Epub 2018 Jan 5. J Glob Antimicrob Resist. 2018. PMID: 29309948
-
Long-lasting outbreak due to CTX-M-15-producing Klebsiella pneumoniae ST336 in a rehabilitation ward: report and literature review.J Hosp Infect. 2017 Sep;97(1):42-51. doi: 10.1016/j.jhin.2017.04.002. Epub 2017 Apr 9. J Hosp Infect. 2017. PMID: 28454769 Review.
Cited by
-
Intestinal colonization with ESBL-producing Klebsiella pneumoniae in healthy rural villager: A genomic surveillance study in China, 2015-2017.Front Public Health. 2022 Dec 15;10:1017050. doi: 10.3389/fpubh.2022.1017050. eCollection 2022. Front Public Health. 2022. PMID: 36589964 Free PMC article.
-
Comparative genomics of an extensively drug resistant strain Klebsiella pneumoniae IITR008 with international high-risk clonal lineage ST147 isolated from river water.Antonie Van Leeuwenhoek. 2024 Mar 15;117(1):57. doi: 10.1007/s10482-024-01955-z. Antonie Van Leeuwenhoek. 2024. PMID: 38491220
-
Emergence of a NDM-1-producing ST25 Klebsiella pneumoniae strain causing neonatal sepsis in China.Front Microbiol. 2022 Oct 20;13:980191. doi: 10.3389/fmicb.2022.980191. eCollection 2022. Front Microbiol. 2022. PMID: 36338063 Free PMC article.
-
Multidrug Resistant Klebsiella pneumoniae ST101 Clone Survival Chain From Inpatients to Hospital Effluent After Chlorine Treatment.Front Microbiol. 2021 Jan 11;11:610296. doi: 10.3389/fmicb.2020.610296. eCollection 2020. Front Microbiol. 2021. PMID: 33584574 Free PMC article.
-
Genomic characterization of two carbapenem-resistant Serratia marcescens isolates causing bacteremia: Emergence of KPC-2-encoding IncR plasmids.Front Cell Infect Microbiol. 2023 Feb 8;13:1075255. doi: 10.3389/fcimb.2023.1075255. eCollection 2023. Front Cell Infect Microbiol. 2023. PMID: 36844412 Free PMC article.
References
-
- An S., Chen J., Wang Z., Wang X., Yan X., Li J., et al. . (2012). Predominant characteristics of CTX-M-producing Klebsiella pneumoniae isolates from patients with lower respiratory tract infection in multiple medical centers in China. FEMS Microbiol. Lett. 332, 137–145. 10.1111/j.1574-6968.2012.02586.x - DOI - PubMed
-
- Avgoulea K., Di Pilato V., Zarkotou O., Sennati S., Politi L., Cannatelli A., et al. . (2018). Characterization of extensively- or pandrug-resistant ST147 and ST101 OXA-48-producing Klebsiella pneumoniae isolates causing bloodstream infections in ICU patients. Antimicrob Agents Chemother. 62, AAC.02457–17. 10.1128/AAC.02457-17 - DOI - PMC - PubMed
-
- Ben Said L., Jouini A., Klibi N., Dziri R., Alonso C. A., Boudabous A., et al. . (2015). Detection of extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in vegetables, soil and water of the farm environment in Tunisia. Int. J. Food Microbiol. 203, 86–92. 10.1016/j.ijfoodmicro.2015.02.023 - DOI - PubMed
LinkOut - more resources
Full Text Sources

