Bacterial 'Grounded' Prophages: Hotspots for Genetic Renovation and Innovation
- PMID: 30809245
- PMCID: PMC6379469
- DOI: 10.3389/fgene.2019.00065
Bacterial 'Grounded' Prophages: Hotspots for Genetic Renovation and Innovation
Abstract
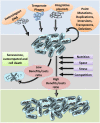
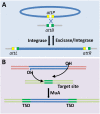
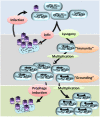
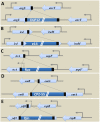
Bacterial genomes are highly plastic allowing the generation of variants through mutations and acquisition of genetic information. The fittest variants are then selected by the econiche thereby allowing the bacterial adaptation and colonization of the habitat. Larger genomes, however, may impose metabolic burden and hence bacterial genomes are optimized by the loss of frivolous genetic information. The activity of temperate bacteriophages has acute consequences on the bacterial population as well as the bacterial genome through lytic and lysogenic cycles. Lysogeny is a selective advantage as the prophage provides immunity to the lysogen against secondary phage attack. Since the non-lysogens are eliminated by the lytic phages, lysogens multiply and colonize the habitat. Nevertheless, all lysogens have an imminent risk of lytic cycle activation and cell lysis. However, a mutation in the attachment sites or in the genes that encode the specific recombinase responsible for prophage excision could result in 'grounding' of the prophage. Since the lysogens with grounded prophage are immune to respective phage infection as well as dodge the induction of lytic cycle, we hypothesize that the selection of these mutant lysogens is favored relative to their normal lysogenic counterparts. These grounded prophages offer several advantages to the bacterial genome evolution through propensity for genetic variations including inversions, deletions, and insertions via horizontal gene transfer. We propose that the grounded prophages expedite bacterial genome evolution by acting as 'genetic buffer zones' thereby increasing the frequency as well as the diversity of variations on which natural selection favors the beneficial variants. The grounded prophages are also hotspots for horizontal gene transfer wherein several ecologically significant genes such as those involved in stress tolerance, antimicrobial resistance, and novel metabolic pathways, are integrated. Moreover, the high frequency of genetic changes within prophages also allows proportionate probability for the de novo genesis of genetic information. Through sequence analyses of well-characterized E. coli prophages we exemplify various roles of grounded prophages in E. coli ecology and evolution. Therefore, the temperate prophages are one of the most significant drivers of bacterial genome evolution and sites of biogenesis of genetic information.
Keywords: bacterial ecology; bacteriophage; genome evolution; genome plasticity; horizontal gene transfer.
Figures

Similar articles
-
An Eco-evolutionary Model on Surviving Lysogeny Through Grounding and Accumulation of Prophages.Microb Ecol. 2023 Nov;86(4):3068-3081. doi: 10.1007/s00248-023-02301-y. Epub 2023 Oct 16. Microb Ecol. 2023. PMID: 37843655
-
Plasmid-Mediated Stabilization of Prophages.mSphere. 2022 Apr 27;7(2):e0093021. doi: 10.1128/msphere.00930-21. Epub 2022 Mar 21. mSphere. 2022. PMID: 35311569 Free PMC article.
-
The book of Lambda does not tell us that naturally occurring lysogens of Escherichia coli are likely to be resistant as well as immune.Proc Natl Acad Sci U S A. 2023 Mar 14;120(11):e2212121120. doi: 10.1073/pnas.2212121120. Epub 2023 Mar 7. Proc Natl Acad Sci U S A. 2023. PMID: 36881631 Free PMC article.
-
Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion.Microbiol Mol Biol Rev. 2004 Sep;68(3):560-602, table of contents. doi: 10.1128/MMBR.68.3.560-602.2004. Microbiol Mol Biol Rev. 2004. PMID: 15353570 Free PMC article. Review.
-
An overview on Vibrio temperate phages: Integration mechanisms, pathogenicity, and lysogeny regulation.Microb Pathog. 2022 Apr;165:105490. doi: 10.1016/j.micpath.2022.105490. Epub 2022 Mar 17. Microb Pathog. 2022. PMID: 35307601 Review.
Cited by
-
Prophages Present in Acinetobacter pittii Influence Bacterial Virulence, Antibiotic Resistance, and Genomic Rearrangements.Phage (New Rochelle). 2022 Mar 1;3(1):38-49. doi: 10.1089/phage.2021.0014. Epub 2022 Mar 18. Phage (New Rochelle). 2022. PMID: 36161193 Free PMC article.
-
Hybrid genome de novo assembly with methylome analysis of the anaerobic thermophilic subsurface bacterium Thermanaerosceptrum fracticalcis strain DRI-13T.BMC Genomics. 2021 Mar 23;22(1):209. doi: 10.1186/s12864-021-07535-z. BMC Genomics. 2021. PMID: 33757423 Free PMC article.
-
Geography shapes the genomics and antimicrobial resistance of Salmonella enterica Serovar Enteritidis isolated from humans.Sci Rep. 2023 Jan 24;13(1):1331. doi: 10.1038/s41598-022-24150-4. Sci Rep. 2023. PMID: 36693882 Free PMC article.
-
An Eco-evolutionary Model on Surviving Lysogeny Through Grounding and Accumulation of Prophages.Microb Ecol. 2023 Nov;86(4):3068-3081. doi: 10.1007/s00248-023-02301-y. Epub 2023 Oct 16. Microb Ecol. 2023. PMID: 37843655
-
Genomic structural plasticity of rodent-associated Bartonella in nature.Mol Ecol. 2022 Jul;31(14):3784-3797. doi: 10.1111/mec.16547. Epub 2022 Jun 10. Mol Ecol. 2022. PMID: 35620948 Free PMC article.
References
-
- Asadulghani M., Ogura Y., Ooka T., Itoh T., Sawaguchi A., Iguchi A., et al. (2009). The defective prophage pool of Escherichia coli O157: prophage-prophage interactions potentiate horizontal transfer of virulence determinants. PLoS Pathog. 5:e1000408. 10.1371/journal.ppat.1000408 - DOI - PMC - PubMed
-
- Bartels M. D., Boye K., Rohde S. M., Larsen A. R., Torfs H., Bouchy P., et al. (2009). A common variant of staphylococcal cassette chromosome mec type IVa in isolates from Copenhagen, Denmark, is not detected by the BD GeneOhm methicillin-resistant Staphylococcus aureus assay. J. Clin. Microbiol. 47 1524–1527. 10.1128/JCM.02153-08 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

