Microscopy-Based Automated Live Cell Screening for Small Molecules That Affect Ciliation
- PMID: 30809247
- PMCID: PMC6379280
- DOI: 10.3389/fgene.2019.00075
Microscopy-Based Automated Live Cell Screening for Small Molecules That Affect Ciliation
Abstract
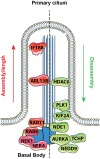
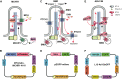
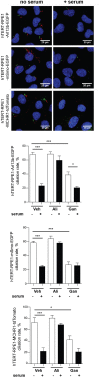
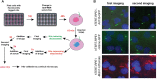
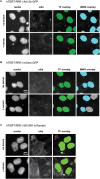
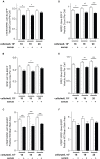
The primary monocilium, or cilium, is a single antenna-like organelle that protrudes from the surface of most mammalian cell types, and serves as a signaling hub. Mutations of cilia-associated genes result in severe genetic disorders termed ciliopathies. Among these, the most common is autosomal dominant polycystic kidney disease (ADPKD); less common genetic diseases include Bardet-Biedl syndrome, Joubert syndrome, nephronophthisis, and others. Important signaling cascades with receptor systems localized exclusively or in part at cilia include Sonic Hedgehog (SHH), platelet derived growth factor alpha (PDGFRα), WNTs, polycystins, and others. Changes in ciliation during development or in pathological conditions such as cancer impacts signaling by these proteins. Notably, ciliation status of cells is coupled closely to the cell cycle, with cilia protruding in quiescent (G0) or early G1 cells, declining in S/G2, and absent in M phase, and has been proposed to contribute to cell cycle regulation. Because of this complex biology, the elaborate machinery regulating ciliary assembly and disassembly receives input from many cellular proteins relevant to cell cycle control, development, and oncogenic transformation, making study of genetic factors and drugs influencing ciliation of high interest. One of the most effective tools to investigate the dynamics of the cilia under different conditions is the imaging of live cells. However, developing assays to observe the primary cilium in real time can be challenging, and requires a consideration of multiple details related to the cilia biology. With the dual goals of identifying small molecules that may have beneficial activity through action on human diseases, and of identifying ciliary activities of existing agents that are in common use or development, we here describe creation and evaluation of three autofluorescent cell lines derived from the immortalized retinal pigmented epithelium parental cell line hTERT-RPE1. These cell lines stably express the ciliary-targeted fluorescent proteins L13-Arl13bGFP, pEGFP-mSmo, and tdTomato-MCHR1-N-10. We then describe methods for use of these cell lines in high throughput screening of libraries of small molecule compounds to identify positive and negative regulators of ciliary disassembly.
Keywords: ADPKD; aurora kinase A; ciliary disassembly; drugs; heat shock protein 90; high content imaging; screening; targeted therapy.
Figures

References
-
- Buchser W., Collins M., Garyantes T., Guha R., Haney S., Lemmon V., et al. (2012). “Assay development guidelines for image-based high content screening, high content analysis and high content imaging,” in Assay Guidance Manual, eds Sittampalam G. S., Coussens N. P., Brimacombe K., Grossman A., Arkin M., Auld D., et al. (Rockville, MD: Bethesda; ).
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

