Nitric oxide increases biofilm formation in Saccharomyces cerevisiae by activating the transcriptional factor Mac1p and thereby regulating the transmembrane protein Ctr1
- PMID: 30809273
- PMCID: PMC6375214
- DOI: 10.1186/s13068-019-1359-1
Nitric oxide increases biofilm formation in Saccharomyces cerevisiae by activating the transcriptional factor Mac1p and thereby regulating the transmembrane protein Ctr1
Abstract
Background: Biofilms with immobilized cells encased in extracellular polymeric substance are beneficial for industrial fermentation. Their formation is regulated by various factors, including nitric oxide (NO), which is recognized as a quorum-sensing and signal molecule. The mechanisms by which NO regulates bacterial biofilms have been studied extensively and deeply, but were rarely studied in fungi. In this study, we observed the effects of low concentrations of NO on biofilm formation in Saccharomyces cerevisiae. Transcriptional and proteomic analyses were applied to study the mechanism of this regulation.
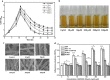
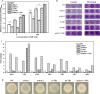
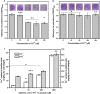
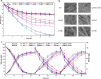
Results: Adding low concentrations of NO donors (SNP and NOC-18) enhanced biofilm formation of S. cerevisiae in immobilized carriers and plastics. Transcriptional and proteomic analyses revealed that expression levels of genes regulated by the transcription factor Mac1p was upregulated in biofilm cells under NO treatment. MAC1 promoted yeast biofilm formation which was independent of flocculation gene FLO11. Increased copper and iron contents, both of which were controlled by Mac1p in the NO-treated and MAC1-overexpressing cells, were not responsible for the increased biofilm formation. CTR1, one out of six genes regulated by MAC1, plays an important role in biofilm formation. Moreover, MAC1 and CTR1 contributed to the cells' resistance to ethanol by enhanced biofilm formation.
Conclusions: These findings suggest that a mechanism for NO-mediated biofilm formation, which involves the regulation of CTR1 expression levels by activating its transcription factor Mac1p, leads to enhanced biofilm formation. The role of CTR1 protein in yeast biofilm formation may be due to the hydrophobic residues in its N-terminal extracellular domain, and further research is needed. This work offers a possible explanation for yeast biofilm formation regulated by NO and provides approaches controlling biofilm formation in industrial immobilized fermentation by manipulating expression of genes involved in biofilm formation.
Keywords: Biofilm; CTR1; MAC1; Nitric oxide; Saccharomyces cerevisiae.
Figures

Similar articles
-
Structure-function analysis of the protein-binding domains of Mac1p, a copper-dependent transcriptional activator of copper uptake in Saccharomyces cerevisiae.J Biol Chem. 1999 Oct 8;274(41):29211-9. doi: 10.1074/jbc.274.41.29211. J Biol Chem. 1999. PMID: 10506178
-
Copper-mediated repression of the activation domain in the yeast Mac1p transcription factor.Proc Natl Acad Sci U S A. 1997 May 27;94(11):5550-5. doi: 10.1073/pnas.94.11.5550. Proc Natl Acad Sci U S A. 1997. PMID: 9159110 Free PMC article.
-
FLO Genes Family and Transcription Factor MIG1 Regulate Saccharomyces cerevisiae Biofilm Formation During Immobilized Fermentation.Front Microbiol. 2018 Aug 23;9:1860. doi: 10.3389/fmicb.2018.01860. eCollection 2018. Front Microbiol. 2018. PMID: 30210459 Free PMC article.
-
Saccharomyces cerevisiae--a model to uncover molecular mechanisms for yeast biofilm biology.FEMS Immunol Med Microbiol. 2012 Jul;65(2):169-82. doi: 10.1111/j.1574-695X.2012.00943.x. Epub 2012 Mar 8. FEMS Immunol Med Microbiol. 2012. PMID: 22332975 Review.
-
Copper-regulatory domain involved in gene expression.Prog Nucleic Acid Res Mol Biol. 1998;58:165-95. doi: 10.1016/s0079-6603(08)60036-7. Prog Nucleic Acid Res Mol Biol. 1998. PMID: 9308366 Review.
Cited by
-
Effect of quorum-sensing molecule 2-phenylethanol and ARO genes on Saccharomyces cerevisiae biofilm.Appl Microbiol Biotechnol. 2021 May;105(9):3635-3648. doi: 10.1007/s00253-021-11280-4. Epub 2021 Apr 14. Appl Microbiol Biotechnol. 2021. PMID: 33852023
-
Quorum sensing: cell-to-cell communication in Saccharomyces cerevisiae.Front Microbiol. 2023 Nov 23;14:1250151. doi: 10.3389/fmicb.2023.1250151. eCollection 2023. Front Microbiol. 2023. PMID: 38075875 Free PMC article. Review.
-
Blue Light Signaling Regulates Escherichia coli W1688 Biofilm Formation and l-Threonine Production.Microbiol Spectr. 2022 Oct 26;10(5):e0246022. doi: 10.1128/spectrum.02460-22. Epub 2022 Sep 27. Microbiol Spectr. 2022. PMID: 36165805 Free PMC article.
-
Calcineurin signaling pathway influences Aspergillus niger biofilm formation by affecting hydrophobicity and cell wall integrity.Biotechnol Biofuels. 2020 Mar 16;13:54. doi: 10.1186/s13068-020-01692-1. eCollection 2020. Biotechnol Biofuels. 2020. PMID: 32190119 Free PMC article.
-
Cell Cycle Progression Influences Biofilm Formation in Saccharomyces cerevisiae 1308.Microbiol Spectr. 2022 Jun 29;10(3):e0276521. doi: 10.1128/spectrum.02765-21. Epub 2022 Jun 7. Microbiol Spectr. 2022. PMID: 35670600 Free PMC article.
References
-
- Blankenship JR, Mitchell AP. How to build a biofilm: a fungal perspective. Curr Opin Microbiol. 2006;9(6):588–594. - PubMed
-
- Liu S, Gunawan C, Barraud N, Rice SA, Harry EJ, Amal R. Understanding, monitoring, and controlling biofilm growth in drinking water distribution systems. Environ Sci Technol. 2016;50(17):8954–8976. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

