Circular RNAs in immune responses and immune diseases
- PMID: 30809295
- PMCID: PMC6376182
- DOI: 10.7150/thno.29678
Circular RNAs in immune responses and immune diseases
Abstract
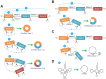
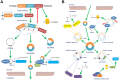
Circular RNAs (circRNAs) are novel clusters of endogenous noncoding RNAs (ncRNAs) that are widely expressed in eukaryotic cells. In contrast to the generation of linear RNA transcripts, circRNAs undergo a "back-splicing" process to form a continuous, covalently closed, stable loop structure without 5' or 3' polarities and poly (A) tails during posttranscriptional modification. Due to the widespread availability of several technologies, especially high-throughput RNA sequencing, numerous circRNAs have been discovered not only in mammals but also in plants and insects. Notably, due to their abilities to serve as microRNA (miRNA) "sponges", miRNA "reservoirs", regulate gene expression and encode proteins, circRNAs participate in the development and progression of different immune responses and immune diseases by enriching various forms of epigenetic modification. CircRNAs have been demonstrated to be expressed in a tissue-specific and pathogenesis-related manner during the occurrence of multiple immune diseases. Additionally, because of their circular configurations, expression in blood and peripheral tissues and coexistence with exosomes, circRNAs show inherent conservation along with environmental resistance stability and may be regarded as potential biomarkers or therapeutic targets for some immune diseases. In this review, we summarize the characteristics, functions and mechanisms of circRNAs and their involvement in immune responses and diseases. Although our knowledge of circRNAs remains preliminary, this field is worthy of deeper exploration and greater research efforts.
Keywords: biogenesis; circRNAs; function; immune diseases; immune responses.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures
References
-
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J. et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24:437–40. - PubMed
-
- Franz A, Rabien A, Stephan C, Ralla B, Fuchs S, Jung K. et al. Circular RNAs: a new class of biomarkers as a rising interest in laboratory medicine. Clin Chem Lab Med. 2018;56:1992–2003. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

