Expression analysis of four pseudo-response regulator (PRR) genes in Chrysanthemum morifolium under different photoperiods
- PMID: 30809439
- PMCID: PMC6385685
- DOI: 10.7717/peerj.6420
Expression analysis of four pseudo-response regulator (PRR) genes in Chrysanthemum morifolium under different photoperiods
Abstract
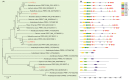
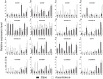
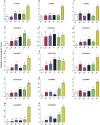
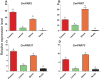
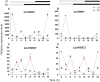
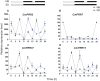
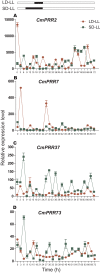
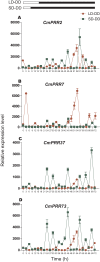
Genes encoding pseudo-response regulator (PRR) proteins play significant roles in plant circadian clocks. In this study, four genes related to flowering time were isolated from Chrysanthemum morifolium. Phylogenetic analysis showed that they are highly homologous to the counterparts of PRRs of Helianthus annuus and named as CmPRR2, CmPRR7, CmPRR37, and CmPRR73. Conserved motifs prediction indicated that most of the closely related members in the phylogenetic tree share common protein sequence motifs, suggesting functional similarities among the PRR proteins within the same subtree. In order to explore functions of the genes, we selected two Chrysanthemum varieties for comparison; that is, a short-day sensitive Zijiao and a short-day insensitive Aoyunbaixue. Compared to Aoyunbaixue, Zijiao needs 13 more days to complete the flower bud differentiation. Evidence from spatio-temporal gene expression patterns demonstrated that the CmPRRs are highly expressed in flower and stem tissues, with a growing trend across the Chrysanthemum developmental process. In addition, we also characterized the CmPRRs expression patterns and found that CmPRRs can maintain their circadian oscillation features to some extent under different photoperiod treatment conditions. These lines of evidence indicated that the four CmPRRs undergo circadian oscillation and possibly play roles in regulating the flowering time of C. morifolium.
Keywords: Chrysanthemum; Circadian clock; Flower bud differentiation; Gene expression; PRR.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Conservation of Arabidopsis thaliana circadian clock genes in Chrysanthemum lavandulifolium.Plant Physiol Biochem. 2014 Jul;80:337-47. doi: 10.1016/j.plaphy.2014.04.001. Epub 2014 Apr 18. Plant Physiol Biochem. 2014. PMID: 24844451
-
Gene cloning, expression pattern analysis, and subcellular localization of LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) homologs in chrysanthemum (Chrysanthemum morifolium Ramat.).Cell Mol Biol (Noisy-le-grand). 2019 Mar 31;65(3):25-31. Cell Mol Biol (Noisy-le-grand). 2019. PMID: 30942153
-
Photoperiodic control of FT-like gene ClFT initiates flowering in Chrysanthemum lavandulifolium.Plant Physiol Biochem. 2014 Jan;74:230-8. doi: 10.1016/j.plaphy.2013.11.004. Epub 2013 Nov 15. Plant Physiol Biochem. 2014. PMID: 24316581
-
Characterization of TEMINAL FLOWER1 homologs CmTFL1c gene from Chrysanthemum morifolium.Plant Mol Biol. 2019 Apr;99(6):587-601. doi: 10.1007/s11103-019-00838-6. Epub 2019 Feb 14. Plant Mol Biol. 2019. PMID: 30762161
-
Transcriptome analysis of the molecular mechanism of Chrysanthemum flower color change under short-day photoperiods.Plant Physiol Biochem. 2020 Jan;146:315-328. doi: 10.1016/j.plaphy.2019.11.027. Epub 2019 Nov 23. Plant Physiol Biochem. 2020. PMID: 31785518
Cited by
-
Genome-wide analysis of the CCT gene family in Chinese white pear (Pyrus bretschneideri Rehd.) and characterization of PbPRR2 in response to varying light signals.BMC Plant Biol. 2022 Feb 23;22(1):81. doi: 10.1186/s12870-022-03476-1. BMC Plant Biol. 2022. PMID: 35196984 Free PMC article.
-
Molecular characterization of PSEUDO RESPONSE REGULATOR family in Rosaceae and function of PbPRR59a and PbPRR59b in flowering regulation.BMC Genomics. 2024 Aug 22;25(1):794. doi: 10.1186/s12864-024-10720-5. BMC Genomics. 2024. PMID: 39169310 Free PMC article.
References
-
- Akinori M, Masakazu K, Yuko N, Takahiko K, Masaya M, Takafumi Y, Takeshi M. Characterization of circadian-associated pseudo-response regulators: II. The function of PRR5 and its molecular dissection in Arabidopsis thaliana. Bioscience Biotechnology and Biochemistry. 2007;71(2):535–544. doi: 10.1271/bbb.60584. - DOI - PubMed
-
- Ariadne MP, Briana DD, Dimitrios G, Maria B, Stavroula B, Dimitrios H. Blood visfatin concentrations in normal full-term pregnancies. Acta Paediatrica. 2002;96:833–843. - PubMed
LinkOut - more resources
Full Text Sources

