Automated Multiscale Approach To Predict Self-Diffusion from a Potential Energy Field
- PMID: 30811190
- PMCID: PMC6460401
- DOI: 10.1021/acs.jctc.8b01255
Automated Multiscale Approach To Predict Self-Diffusion from a Potential Energy Field
Abstract
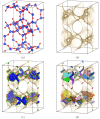
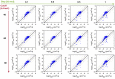
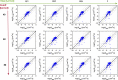
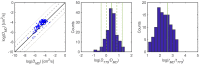
For large-scale screening studies there is a need to estimate the diffusion of gas molecules in nanoporous materials more efficiently than (brute force) molecular dynamics. In particular for systems with low diffusion coefficients molecular dynamics can be prohibitively expensive. An alternative is to compute the hopping rates between adsorption sites using transition state theory. For large-scale screening this requires the automatic detection of the transition states between the adsorption sites along the different diffusion paths. Here an algorithm is presented that analyzes energy grids for the moving particles. It detects the energies at which diffusion paths are formed, together with their directions. This allows for easy identification of nondiffusive systems. For diffusive systems, it partitions the grid coordinates assigned to energy basins and transitions states, permitting a transition state theory based analysis of the diffusion. We test our method on CH4 diffusion in zeolites, using a standard kinetic Monte Carlo simulation based on the output of our grid analysis. We find that it is accurate, fast, and rigorous without limitations to the geometries of the diffusion tunnels or transition states.
Conflict of interest statement
The authors declare no competing financial interest.
Figures







References
-
- Qiao Z.; Xu Q.; Jiang J. High-throughput computational screening of metal-organic framework membranes for upgrading of natural gas. J. Membr. Sci. 2018, 551, 47–54. 10.1016/j.memsci.2018.01.020. - DOI
