β-Galactosidase from Lactobacillus helveticus DSM 20075: Biochemical Characterization and Recombinant Expression for Applications in Dairy Industry
- PMID: 30813223
- PMCID: PMC6412629
- DOI: 10.3390/ijms20040947
β-Galactosidase from Lactobacillus helveticus DSM 20075: Biochemical Characterization and Recombinant Expression for Applications in Dairy Industry
Abstract
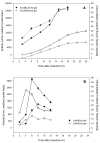
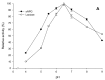
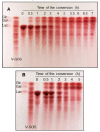
β-Galactosidase encoding genes lacLM from Lactobacillus helveticus DSM 20075 were cloned and successfully overexpressed in Escherichia coli and Lactobacillus plantarum using different expression systems. The highest recombinant β-galactosidase activity of ∼26 kU per L of medium was obtained when using an expression system based on the T7 RNA polymerase promoter in E. coli, which is more than 1000-fold or 28-fold higher than the production of native β-galactosidase from L. helveticus DSM 20075 when grown on glucose or lactose, respectively. The overexpression in L. plantarum using lactobacillal food-grade gene expression system resulted in ∼2.3 kU per L of medium, which is approximately 10-fold lower compared to the expression in E. coli. The recombinant β-galactosidase from L. helveticus overexpressed in E. coli was purified to apparent homogeneity and subsequently characterized. The Km and vmax values for lactose and o-nitrophenyl-β-d-galactopyranoside (oNPG) were 15.7 ± 1.3 mM, 11.1 ± 0.2 µmol D-glucose released per min per mg protein, and 1.4 ± 0.3 mM, 476 ± 66 µmol o-nitrophenol released per min per mg protein, respectively. The enzyme was inhibited by high concentrations of oNPG with Ki,s = 3.6 ± 0.8 mM. The optimum pH for hydrolysis of both substrates, lactose and oNPG, is pH 6.5 and optimum temperatures for these reactions are 60 and 55 °C, respectively. The formation of galacto-oligosaccharides (GOS) in discontinuous mode using both crude recombinant enzyme from L. plantarum and purified recombinant enzyme from E. coli revealed high transgalactosylation activity of β-galactosidases from L. helveticus; hence, this enzyme is an interesting candidate for applications in lactose conversion and GOS formation processes.
Keywords: Lactobacillus helveticus; expression systems; recombinant enzyme; β-galactosidase.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





Similar articles
-
beta-Galactosidase from Lactobacillus plantarum WCFS1: biochemical characterization and formation of prebiotic galacto-oligosaccharides.Carbohydr Res. 2010 Jul 2;345(10):1408-16. doi: 10.1016/j.carres.2010.03.028. Epub 2010 Mar 21. Carbohydr Res. 2010. PMID: 20385377
-
Characterization of a heterodimeric GH2 β-galactosidase from Lactobacillus sakei Lb790 and formation of prebiotic galacto-oligosaccharides.J Agric Food Chem. 2011 Apr 27;59(8):3803-11. doi: 10.1021/jf103832q. Epub 2011 Mar 25. J Agric Food Chem. 2011. PMID: 21405014
-
Construction and Analysis of Food-Grade Lactobacillus kefiranofaciens β-Galactosidase Overexpression System.J Microbiol Biotechnol. 2021 Apr 28;31(4):550-558. doi: 10.4014/jmb.2101.01028. J Microbiol Biotechnol. 2021. PMID: 33622994 Free PMC article.
-
Cloning, purification, and characterization of β-galactosidase from Bacillus licheniformis DSM 13.Appl Microbiol Biotechnol. 2011 Feb;89(3):645-54. doi: 10.1007/s00253-010-2862-2. Epub 2010 Sep 18. Appl Microbiol Biotechnol. 2011. PMID: 20852995
-
Invited review: Properties of β-galactosidases derived from Lactobacillaceae species and their capacity for galacto-oligosaccharide production.J Dairy Sci. 2023 Dec;106(12):8193-8206. doi: 10.3168/jds.2023-23392. Epub 2023 Sep 9. J Dairy Sci. 2023. PMID: 37678769 Review.
Cited by
-
A Review on the Various Sources of β-Galactosidase and Its Lactose Hydrolysis Property.Curr Microbiol. 2023 Mar 2;80(4):122. doi: 10.1007/s00284-023-03220-4. Curr Microbiol. 2023. PMID: 36862237 Review.
-
Isolation and characterization of a β-galactosidase from Lactobacillus helveticus for industrial processing.JDS Commun. 2024 Jun 13;6(1):19-23. doi: 10.3168/jdsc.2024-0563. eCollection 2025 Jan. JDS Commun. 2024. PMID: 39877184 Free PMC article.
-
Genomic insights into habitat adaptation of Lactobacillus species.World J Microbiol Biotechnol. 2025 Feb 4;41(2):61. doi: 10.1007/s11274-025-04275-0. World J Microbiol Biotechnol. 2025. PMID: 39900839 Free PMC article.
-
Immobilization of β-Galactosidases on the Lactobacillus Cell Surface Using the Peptidoglycan-Binding Motif LysM.Catalysts. 2019 May;9(5):443. doi: 10.3390/catal9050443. Catalysts. 2019. PMID: 31595189 Free PMC article.
-
A Comparative Genomic and Safety Assessment of Six Lactiplantibacillus plantarum subsp. argentoratensis Strains Isolated from Spontaneously Fermented Greek Wheat Sourdoughs for Potential Biotechnological Application.Int J Mol Sci. 2022 Feb 24;23(5):2487. doi: 10.3390/ijms23052487. Int J Mol Sci. 2022. PMID: 35269627 Free PMC article.
References
-
- Ljungh A., Wadström T. Lactic acid bacteria as probiotics. Curr. Issues Intest. Microbiol. 2006;7:73–89. - PubMed
-
- Gatti M., Trivisano C., Fabrizi E., Neviani E., Gardini F. Biodiversity among Lactobacillus helveticus strains isolated from different natural whey starter cultures as revealed by classification trees. Appl. Environ. Microbiol. 2004;70:182–190. doi: 10.1128/AEM.70.1.182-190.2004. - DOI - PMC - PubMed
-
- de Macías M.E., Manca de Nadra M.C., Strasser de Saad A.M., Pesce de Ruiz Holgado A.A., Oliver G. Isolation and properties of β-galactosidase of a strain of Lactobacillus helveticus isolated from natural whey starter. J. Appl. Biochem. 1983;5:275–281. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

