Myc-like transcriptional factors in wheat: structural and functional organization of the subfamily I members
- PMID: 30813892
- PMCID: PMC6393960
- DOI: 10.1186/s12870-019-1639-8
Myc-like transcriptional factors in wheat: structural and functional organization of the subfamily I members
Abstract
Background: Myc-like regulatory factors carrying the basic helix-loop-helix (bHLH) domain belong to a large superfamily of transcriptional factors (TFs) present in all eukaryotic kingdoms. In plants, the representatives of this superfamily regulate diverse biological processes including growth and development as well as response to various stresses. As members of the regulatory MBW complexes, they participate in biosynthesis of flavonoids. In wheat, only one member (TaMyc1) of the Myc-like TFs family has been studied, while structural and functional organization of further members remained uncharacterized. From two Myc-subfamilies described recently in the genomes of Triticeae tribe species, we investigated thoroughly the members of the subfamily I which includes the TaMyc1 gene.
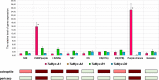
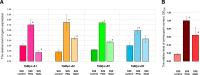
Results: Comparison of the promoter regions of the Myc subfamily I members in wheat suggested their division into two groups (likely homoeologous sets): TaMyc-1 (TaMyc-A1/TaMyc1, TaMyc-B1, TaMyc-D1) and TaMyc-2 (TaMyc-A2 and TaMyc-D2). It was demonstrated that the TaMyc-D1 copy has lost its functionality due to the frame shift mutation. The study of functional features of the other four copies suggested some of them to be involved in the biosynthesis of anthocyanins. In particular, TaMyc-B1 is assumed to be a co-regulator of the gene TaC1-A1 (encoding R2R3-Myb factor) in the MBW regulatory complex activating anthocyanin synthesis in wheat coleoptile. The mRNA levels of the TaMyc-A1, TaMyc-B1, TaMyc-A2 and TaMyc-D2 genes increased significantly in wheat seedlings exposed to osmotic stress. Salinity stress induced expression of TaMyc-B1 and TaMyc-A2, while TaMyc-A1 was repressed.
Conclusions: The features of the structural and functional organization of the members of subfamily I of Myc-like TFs in wheat were determined. Myc-like co-regulator (TaMyc-B1) of anthocyanin synthesis in wheat coleoptile was described for the first time. The Myc-encoding genes presumably involved in response to drought and salinity were determined in wheat. The results obtained are important for further manipulations with Myc genes, aimed on increasing wheat adaptability.
Keywords: Anthocyanin biosynthesis; Flavonoid biosynthesis; Gene duplication; Myc; Osmotic stress; Salinity stress; Stress response; Transcription factor; Triticum; Wheat; bHLH.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures





References
-
- Cooper GM. The cell: a molecular approach. 2nd edition. Sunderland: Sinauer Associates; 2000. Regulation of transcription in eukaryotes.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

