The pluripotency factor NANOG controls primitive hematopoiesis and directly regulates Tal1
- PMID: 30814124
- PMCID: PMC6443201
- DOI: 10.15252/embj.201899122
The pluripotency factor NANOG controls primitive hematopoiesis and directly regulates Tal1
Abstract
Progenitors of the first hematopoietic cells in the mouse arise in the early embryo from Brachyury-positive multipotent cells in the posterior-proximal region of the epiblast, but the mechanisms that specify primitive blood cells are still largely unknown. Pluripotency factors maintain uncommitted cells of the blastocyst and embryonic stem cells in the pluripotent state. However, little is known about the role played by these factors during later development, despite being expressed in the postimplantation epiblast. Using a dual transgene system for controlled expression at postimplantation stages, we found that Nanog blocks primitive hematopoiesis in the gastrulating embryo, resulting in a loss of red blood cells and downregulation of erythropoietic genes. Accordingly, Nanog-deficient embryonic stem cells are prone to erythropoietic differentiation. Moreover, Nanog expression in adults prevents the maturation of erythroid cells. By analysis of previous data for NANOG binding during stem cell differentiation and CRISPR/Cas9 genome editing, we found that Tal1 is a direct NANOG target. Our results show that Nanog regulates primitive hematopoiesis by directly repressing critical erythroid lineage specifiers.
Keywords: Nanog; Tal1; erythropoiesis; gastrulation; primitive hematopoiesis.
© 2019 The Authors. Published under the terms of the CC BY 4.0 license.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
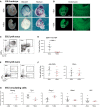
Dox‐induced prolongation of Nanog expression in Nanog tg embryos up to E9.5 results in lack of blood (left) and downregulation of erythropoietic gene expression. The center and right panels show whole‐mount in situ hybridization for Hbb‐bh1 (in embryos with intact yolk sacs) and for the long non‐coding RNA Redrum. Asterisks mark the aorta‐gonad‐mesonephros (AGM) region and arrows the tail bud. Embryos of the same genotype but not treated with dox were used as controls (−dox). Scale bars, 500 μm.
Endomucin staining of vessels in control (−dox) or treated (+dox) E9.5 Nanog tg embryos. On the right, higher magnifications of the boxed areas. Scale bar, 500 μm.
Representative FACS plot of the distribution of the CD71 and Ter119 populations in dissected yolk sacs from untreated and dox‐treated E9.5 Nanog tg embryos.
Quantification of the CD71+ Ter119+ population in controls (−dox, black dots; n = 8) and Nanog expressing (+dox, red dots; n = 7) E9.5 yolk sacs. Each replicate contained a pool of 5 (−dox) or 8 (+dox) E9.5 Nanog tg embryos. ***P < 0.0005; Student's t‐test. Horizontal line represents mean values and error bars standard deviation (SD).
Representative FACS plots showing the distribution of cKit and CD41 populations in yolk sacs from untreated controls (−dox) and Nanog expressing (+dox) E9.5 Nanog tg embryos.
Quantification of different progenitor populations in yolk sacs from control (−dox, black dots; n = 8) and Nanog expressing (+dox, red dots; n = 7) E9.5 embryos. Each replicate contained a pool of 5 (−dox) or 8 (+dox) E9.5 Nanog tg embryos. Horizontal line represents mean values and error bars SD.
Differences in the expression levels of Nanog and selected hematopoietic genes in the CD71+ Ter119+ population of control (−dox; n = 7) and Nanog expressing (+dox; n = 4) E9.5 embryos. **P < 0.005, ***P < 0.0005; Student's t‐test. Horizontal line represents mean values and error bars SD.

CD31 staining of yolk sac vasculature in control (−dox) or treated (+dox) E9.5 Nanog tg embryos. Below, higher magnifications of the boxed areas are shown. Scale bar, 500 μm.
Heart morphology is not affected in dox‐treated (+dox) E9.5 Nanog tg embryos. Below, hematoxylin eosin staining of sections reveal normal development of the heart in treated (+dox). Dotted lines in upper panels indicate plane of sections. Scale bar, 500 μm (whole mounts), 250 μm (sections).
Representative images of May‐Grünwald‐Giemsa stained cytospins from control (−dox) and dox‐treated (+dox) E9.5 embryos. Scale bar, 5 μm.
Relative expression of Nanog and hematopoietic genes in cKit+CD41+ and cKit−CD41+ populations sorted from E9.5 control (−dox) and treated (+dox) embryos. n = 7 (−dox) or n = 4 (+dox); each replicate contained a pool of 5 (−dox) or 8 (+dox) E9.5 Nanog tg embryos. ***P < 0.0005; Student's t‐test. Horizontal line represents mean values and error bars SD.
Whole‐mount in situ hybridization for Gata1 and Klf1 of control (−dox) and treated (+dox) E7.5 Nanog tg embryos. Arrows indicate the location of blood islands in the extraembryonic yolk sac. Scale bar, 250 μm.
Relative expression of Nanog, mesodermal (Eomes, Brachyury, Kdr), and hematopoietic (Runx1, Tal1, Gata1, Klf1) genes in single control (−dox) or treated (+dox) E7.5 embryos (n = 4). *P < 0.05, **P < 0.005, ***P < 0.0005; Student's t‐test. Horizontal line represents mean values and error bars SD.

- A
Experimental design for chimera generation, and contribution of GFP cells to chimeric embryo (right hand side panels).
- B
Freshly dissected dox‐treated Nanog tg E10.5 embryos without (control) and with (chimera) contribution of wt‐ESGFP cells (left, brightfield; right, GFP). Arrows mark the presence of blood in chimeric embryos that is absent from controls. Scale bar, 500 μm.
- C
Representative FACS plots showing of red blood cell maturation as determined by CD71/Ter119 staining in single dox‐treated E10.5 control (left) and chimeric (right) embryos.
- D–F
Quantification of the CD71+ Ter119+ population in single dox‐treated E10.5 embryos (D; control, n = 18; chimera, n = 10), untreated embryos (E; control, n = 7; chimera, n = 12), and in GFP− cells (not derived from wild‐type ES cells) from dox‐treated embryos (F; control, n = 18; chimera, n = 10). ***P < 0.0005; Student's t‐test. Horizontal line represents mean values and error bars SD.

Quantification of colony‐forming units generated by wild‐type (wt) and knockout (Nanog −/−) ES cells after culture of EBs for 5 (D5), 6 (D6), or 7 (D7) days and plating disaggregated cells in different hemogenic‐promoting conditions. Panels on the left show representative images of mouse hematopoietic colonies obtained after 12 days of culture in specific media. CFU‐GEMM, progenitors giving rise to granulocytes, erythrocytes, monocytes, and megakaryocytes; BFU‐E, burst forming units–erythroid; Ery‐P, colony‐forming primitive erythroid; CFU‐GM, granulocyte–monocyte precursors; CFU‐M, monocyte precursors; CFU‐G, granulocyte precursors. No CFU‐GEMM are detected at D5 and no BFU‐E at D7. For both wt and knockout cells, n = 3 each with three technical replicates. *P < 0.05, **P < 0.005, ***P < 0.00005; Student's t‐test. Horizontal line represents mean values and error bars SD.
RT–qPCR determination of the relative expression of Brachyury and selected hematopoietic genes in control (wt, right) and knockout (Nanog −/−, left) ES cells (n = 3) during 10 days of EB differentiation in hematopoietic cytokine‐enriched medium. Black arrowheads indicate the peak of Brachyury expression and white arrowheads the time of maximum hematopoietic gene expression.

Timing of Brachyury expression before (left) and after (right) the alignment of its peak of expression that occurs at day 3 (D3) of differentiation in wild‐type ES cells (wt, black) and at day 5 (D5) in Nanog −/− cells (red). n = 3. Horizontal line represents mean values and error bars SEM.
Timing of expression of Nanog and selected hematopoietic genes when wt and Nanog −/− cells after alignment. The time point of maximum Brachyury expression is labeled as T d0. Horizontal line represents mean values and error bars SEM.
Relative expression of Nanog, Brachyury, Tal1, Gata1, Klf1, and Hbb‐bh1 determined by RT–qPCR for Nanog flox/− and Nanog del/− during ES to EpiL cell transition (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; ANOVA with Fisher post‐test. Horizontal line represents mean values and error bars SEM.

Experimental design for the treatment of adult Nanog tg mice.
Representative FACS plots showing the distribution of different populations distinguished by CD71/Ter119 staining in whole bone marrow from untreated (−dox) or treated (+dox) adult mice. S0 (double negative cell), S1 (proerythroblast), S2 (basophilic erythroblast), S3 (polychromatic erythroblast), and S4 (orthochromatic erythroblast) are different stages of blood maturation.
Quantification of the S1–S4 erythroid populations (−dox, n = 4; +dox, n = 5). *P < 0.05, **P < 0.005, ***P < 0.0005; Student's t‐test. Horizontal line represents mean values and error bars SD.
Representative FACS plots showing the distribution of CD16/32 and CD34 hematopoietic precursors sorted from the cKit+Sca1−LIN− bone marrow of untreated (−dox) or treated (+dox) adult Nanog tg mice.
Quantification of precursor populations based on CD16/32 and CD34 sorting, as total number of cells per individual femur (−dox, n = 5; +dox, n = 6). *P < 0.05, **P < 0.005; Student's t‐test. Horizontal line represents mean values and error bars SD.
RT–qPCR quantification of the relative expression of hematopoietic genes in megakaryocyte–erythroid progenitors (MEP; −dox, n = 8; +dox, n = 5). *P < 0.05, ****P < 0.00005; Student's t‐test. Horizontal line represents mean values and error bars SD.
Experimental design for the transplant of Nanog tg bone marrow to wild‐type recipients and treatment of chimeric mice.
Contribution of Nanog tg transplanted bone marrow cells to peripheral blood before (left) and after (right) dox treatment. Percentage of host‐derived cells (CD45.1+) are shown in black, and of donor derived cells (CD45.1/CD45.2 double +) in red. Individual mice are indicated on the x‐axis (n = 7).
Contribution of Nanog tg transplanted cells to LSK, CMP, GMP, and MEP populations purified from bone marrow. Percentage of host‐derived cells (CD45.1+) are show in black, and of donor derived cells (CD45.1/CD45.2 double +) in blue. Individual mice are indicated on the x‐axis (n = 7).
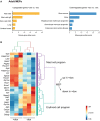
Enriched functional categories in genes that are significantly upregulated (left, orange) or downregulated (right, blue) in MEPs isolated from dox‐treated Nanog tg mice compared to untreated controls. Mouse gene atlas score was calculated using Enrichr.
Heatmap of the expression (as z‐score) across the three replicates for each condition of selected genes for the mast cell and erythroid transcriptional programs.
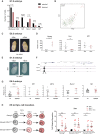
Expected and observed number of mesodermal (Flk1+) cells of the E7.0 mouse embryo expressing Nanog and selected mesodermal or hematopoietic gene expression, based on single‐cell RNA‐seq data (Scialdone et al, 2016). Statistical significance was calculated with a hypergeometric test.
PCA showing the distribution of Flk1+ E7.0 mesoderm cells expressing Nanog (green) or Tal1 (red). The single cell expressing both genes is shown in yellow and indicated by an arrow.
E6.5 Nanog tg embryos after 8 h ex utero culture in the presence (+dox) or absence (−dox) of doxycycline. Scale bar, 100 μm.
RT–qPCR quantification of the relative expression of Nanog, Tal1, and Klf1 in individual untreated embryos (−dox) or treated embryos (+dox) (n = 5). **P < 0.005, ***P < 0.0005; Student's t‐test. Horizontal line represents mean values and error bars SD.
Whole‐mount in situ hybridization of Tal1 in E7.5 untreated (−dox) or in utero treated (+dox) Nanog tg embryos. Scale bar, 100 μm.
UCSC browser view of the Tal1/Stil1 region (mm9; chr4:114,705,753‐114,756,741), indicating the presence of the NANOG binding peak, determined by ChIP‐seq, in EpiLCs (2 replicates are shown) but not in ES cells (Murakami et al, 2016); the binding peak was deleted by CRISPR/Cas9 genome editing (scissors).
RT–qPCR determination of relative expression in wild‐type and CRISPR‐deleted embryos (n = 5) of Tal1 (wt, n = 19; deleted, n = 13), Klf1 (wt, n = 3; deleted, n = 6), Gfi1b (wt, n = 10; deleted, n = 8), Runx1 (wt, n = 13; deleted, n = 5), and Stil (wt, n = 19; deleted, n = 13). **P < 0.005, Student's t‐test. Horizontal line represents mean values and error bars SD.
Experimental design for ES to EpiL cell differentiation of Nanog tg cells and two independent clones (Nanog tg ;dTal1 del#1 and Nanog tg ;dTal1 del#2) where the binding site for NANOG distal to Tal1 has been deleted (left). On the right, relative expression of Tal1 determined by RT–qPCR for each ES cell line (ESC; n = 9 for all three lines) and EpiL cells without (EpiLC; Nanog tg and Nanog tg ;dTal1 del#1 , n = 8; Nanog tg ;dTal1 del#2 , n = 6) or with dox treatment (EpiLC +dox; n = 9 for all three lines). The genotype of the cell lines is indicated below. Values were normalized to Nanog tg ESC. *P < 0.05, **P < 0.01, ns = not significant; ANOVA with Fisher post‐test. Horizontal line represents mean values and error bars standard error of the mean (SEM).
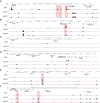
- A–F
UCSC browser views (mm9) of different genomic regions showing NANOG bound regions as determined by ChIP‐seq in ES cells (ESC) and two replicates of epiblast‐like cells (EpiLC1, EpiLC2). Selected peaks are highlighted by boxing in red. (A) Nanog: chr6:122,569,855‐122,699,399. (B) Cdx2: chr5:148,080,850‐148,136,544. (C) Runx1: chr16:92,497,828‐92,940,224. (D) Klf1: chr8:87,395,041‐87,462,459. (E) Tal1: chr4:114,664,868‐114,780,341. (F) Gata1: chrX:7,493,906‐7,601,563. ChIP‐seq data were obtained from Murakami et al, 2016 (GEO accession number GSE71933).
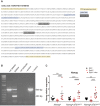
DNA sequence of the genomic region located at −22 kb from Tal1 bound by NANOG in EpiLC. PCR genotyping primers are highlighted in yellow, guide RNAs in blue (PAM sequence is underlined), and two consensus NANOG binding motifs in dark gray and white bold lettering.
Representative gel of PCR genotyping of individual E6.5 embryos showing not deleted, deleted, not detected, negative control (no DNA), and positive control (wild‐type embryo). The size of the wild‐type (949 bp) and deleted (400 bp) bands are indicated.
Relative expression of Nanog determined by RT–qPCR for each ES cell line (ESC; n = 9 for all three lines) and EpiL cells without (EpiLC; EpiLC; Nanog tg and Nanog tg ;dTal1 del#1 , n = 8; Nanog tg ;dTal1 del#2 , n = 7) or with dox treatment (EpiLC +dox; Nanog tg, n = 8; Nanog tg ;dTal1 del#1 and Nanog tg ;dTal1 del#2 , n = 9). The genotype of the cell lines is indicated below. **P < 0.01, ***P < 0.001, ****P < 0.0001; ANOVA with Fisher post‐test. Horizontal line represents mean values and error bars SEM.
References
-
- Acloque H, Wilkinson DG, Nieto MA (2008) In situ hybridization analysis of chick embryos in whole‐mount and tissue sections. Methods Cell Biol 87: 169–185 - PubMed
-
- Aires R, Jurberg AD, Leal F, Novoa A, Cohn MJ, Mallo M (2016) Oct4 is a key regulator of vertebrate trunk length diversity. Dev Cell 38: 262–274 - PubMed
-
- Ariza‐McNaughton L, Krumlauf R (2002) Non‐radioactive in situ hybridization: simplified procedures for use in whole‐mounts of mouse and chick embryos. Int Rev Neurobiol 47: 239–250 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

