EMT is associated with an epigenetic signature of ECM remodeling genes
- PMID: 30814494
- PMCID: PMC6393505
- DOI: 10.1038/s41419-019-1397-4
EMT is associated with an epigenetic signature of ECM remodeling genes
Abstract
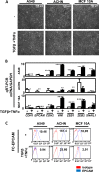
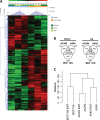
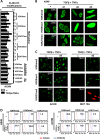
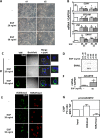
Type III epithelial-mesenchymal transition (EMT) has been previously associated with increased cell migration, invasion, metastasis, and therefore cancer aggressiveness. This reversible process is associated with an important gene expression reprogramming mainly due to epigenetic plasticity. Nevertheless, most of the studies describing the central role of epigenetic modifications during EMT were performed in a single-cell model and using only one mode of EMT induction. In our study, we studied the overall modulations of gene expression and epigenetic modifications in four different EMT-induced cell models issued from different tissues and using different inducers of EMT. Pangenomic analysis (transcriptome and ChIP-sequencing) validated our hypothesis that gene expression reprogramming during EMT is largely regulated by epigenetic modifications of a wide range of genes. Indeed, our results confirmed that each EMT model is unique and can be associated with a specific transcriptome profile and epigenetic program. However, we could select some genes or pathways that are similarly regulated in the different models and that could therefore be used as a common signature of all EMT models and become new biomarkers of the EMT phenotype. As an example, we can cite the regulation of gene-coding proteins involved in the degradation of the extracellular matrix (ECM), which are highly induced in all EMT models. Based on our investigations and results, we identified ADAM19 as a new biomarker of in vitro and in vivo EMT and we validated this biological new marker in a cohort of non-small lung carcinomas.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

