Man-made microbial resistances in built environments
- PMID: 30814504
- PMCID: PMC6393488
- DOI: 10.1038/s41467-019-08864-0
Man-made microbial resistances in built environments
Abstract
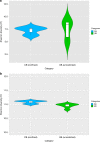
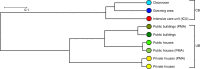
Antimicrobial resistance is a serious threat to global public health, but little is known about the effects of microbial control on the microbiota and its associated resistome. Here we compare the microbiota present on surfaces of clinical settings with other built environments. Using state-of-the-art metagenomics approaches and genome and plasmid reconstruction, we show that increased confinement and cleaning is associated with a loss of microbial diversity and a shift from Gram-positive bacteria, such as Actinobacteria and Firmicutes, to Gram-negative such as Proteobacteria. Moreover, the microbiome of highly maintained built environments has a different resistome when compared to other built environments, as well as a higher diversity in resistance genes. Our results highlight that the loss of microbial diversity correlates with an increase in resistance, and the need for implementing strategies to restore bacterial diversity in certain built environments.
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- O’Neill, J. Tackling drug-resistant infections globally-Final Report and Recommendations. (the Wellcome Trust and the UK Department of Health: United Kingdom 2016).
-
- Bengtsson-Palme, J., Larsson, D. G. J. & Kristiansson, E. Using metagenomics to investigate human and environmental resistomes. J. Antimicrob. Chemother. 10.1093/jac/dkx199 (2017). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

