Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome
- PMID: 30814529
- PMCID: PMC6393466
- DOI: 10.1038/s41467-019-08880-0
Tightly-orchestrated rearrangements govern catalytic center assembly of the ribosome
Abstract
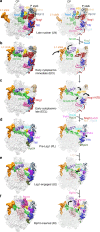
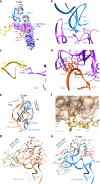
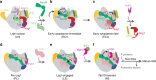
The catalytic activity of the ribosome is mediated by RNA, yet proteins are essential for the function of the peptidyl transferase center (PTC). In eukaryotes, final assembly of the PTC occurs in the cytoplasm by insertion of the ribosomal protein Rpl10 (uL16). We determine structures of six intermediates in late nuclear and cytoplasmic maturation of the large subunit that reveal a tightly-choreographed sequence of protein and RNA rearrangements controlling the insertion of Rpl10. We also determine the structure of the biogenesis factor Yvh1 and show how it promotes assembly of the P stalk, a critical element for recruitment of GTPases that drive translation. Together, our structures provide a blueprint for final assembly of a functional ribosome.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Diverse roles of assembly factors revealed by structures of late nuclear pre-60S ribosomes.Nature. 2016 Jun 2;534(7605):133-7. doi: 10.1038/nature17942. Epub 2016 May 25. Nature. 2016. PMID: 27251291 Free PMC article.
-
Crystal structure of the eukaryotic ribosome.Science. 2010 Nov 26;330(6008):1203-9. doi: 10.1126/science.1194294. Science. 2010. PMID: 21109664
-
Structural snapshot of cytoplasmic pre-60S ribosomal particles bound by Nmd3, Lsg1, Tif6 and Reh1.Nat Struct Mol Biol. 2017 Mar;24(3):214-220. doi: 10.1038/nsmb.3364. Epub 2017 Jan 23. Nat Struct Mol Biol. 2017. PMID: 28112732 Free PMC article.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Ribosome biogenesis in the yeast Saccharomyces cerevisiae.Genetics. 2013 Nov;195(3):643-81. doi: 10.1534/genetics.113.153197. Genetics. 2013. PMID: 24190922 Free PMC article. Review.
Cited by
-
RSL24D1 sustains steady-state ribosome biogenesis and pluripotency translational programs in embryonic stem cells.Nat Commun. 2023 Jan 23;14(1):356. doi: 10.1038/s41467-023-36037-7. Nat Commun. 2023. PMID: 36690642 Free PMC article.
-
RNA folding and functions of RNA helicases in ribosome biogenesis.RNA Biol. 2022 Jan;19(1):781-810. doi: 10.1080/15476286.2022.2079890. RNA Biol. 2022. PMID: 35678541 Free PMC article. Review.
-
Characterization of Single Gene Deletion Mutants Affecting Alternative Oxidase Production in Neurospora crassa: Role of the yvh1 Gene.Microorganisms. 2020 Aug 4;8(8):1186. doi: 10.3390/microorganisms8081186. Microorganisms. 2020. PMID: 32759834 Free PMC article.
-
In situ single particle classification reveals distinct 60S maturation intermediates in cells.Elife. 2022 Aug 25;11:e79272. doi: 10.7554/eLife.79272. Elife. 2022. PMID: 36005291 Free PMC article.
-
The RNA helicase Dbp10 coordinates assembly factor association with PTC maturation during ribosome biogenesis.Nucleic Acids Res. 2024 Feb 28;52(4):1975-1987. doi: 10.1093/nar/gkad1206. Nucleic Acids Res. 2024. PMID: 38113283 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

