An integrative systems genetic analysis of mammalian lipid metabolism
- PMID: 30814737
- PMCID: PMC6656374
- DOI: 10.1038/s41586-019-0984-y
An integrative systems genetic analysis of mammalian lipid metabolism
Abstract
Dysregulation of lipid homeostasis is a precipitating event in the pathogenesis and progression of hepatosteatosis and metabolic syndrome. These conditions are highly prevalent in developed societies and currently have limited options for diagnostic and therapeutic intervention. Here, using a proteomic and lipidomic-wide systems genetic approach, we interrogated lipid regulatory networks in 107 genetically distinct mouse strains to reveal key insights into the control and network structure of mammalian lipid metabolism. These include the identification of plasma lipid signatures that predict pathological lipid abundance in the liver of mice and humans, defining subcellular localization and functionality of lipid-related proteins, and revealing functional protein and genetic variants that are predicted to modulate lipid abundance. Trans-omic analyses using these datasets facilitated the identification and validation of PSMD9 as a previously unknown lipid regulatory protein. Collectively, our study serves as a rich resource for probing mammalian lipid metabolism and provides opportunities for the discovery of therapeutic agents and biomarkers in the setting of hepatic lipotoxicity.
Conflict of interest statement
Figures


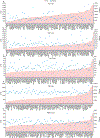






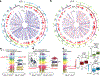
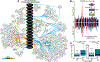

Comment in
-
Understanding lipid metabolism through hepatic steat-omics.Nat Rev Endocrinol. 2019 Jun;15(6):321-322. doi: 10.1038/s41574-019-0203-9. Nat Rev Endocrinol. 2019. PMID: 30979957 No abstract available.
-
Predicting and reducing hepatic lipotoxicity in non-alcoholic fatty liver disease.Lab Anim (NY). 2019 May;48(5):143-144. doi: 10.1038/s41684-019-0291-0. Lab Anim (NY). 2019. PMID: 31000821 No abstract available.
References
-
- Younossi ZM et al. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 64, 73–84 (2016). - PubMed
-
- Sookoian S & Pirola CJ The genetic epidemiology of nonalcoholic fatty liver disease: toward a personalized medicine. Clin. Liver Dis 16, 467–485 (2012). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

