Exposure to Sub-inhibitory Concentrations of the Chemosensitizer 1-(1-Naphthylmethyl)-Piperazine Creates Membrane Destabilization in Multi-Drug Resistant Klebsiella pneumoniae
- PMID: 30814979
- PMCID: PMC6381021
- DOI: 10.3389/fmicb.2019.00092
Exposure to Sub-inhibitory Concentrations of the Chemosensitizer 1-(1-Naphthylmethyl)-Piperazine Creates Membrane Destabilization in Multi-Drug Resistant Klebsiella pneumoniae
Abstract
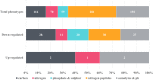
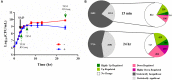
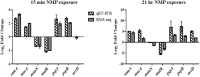
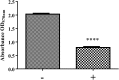
Antimicrobial efflux is one of the important mechanisms causing multi-drug resistance (MDR) in bacteria. Chemosensitizers like 1-(1-naphthylmethyl)-piperazine (NMP) can inhibit an efflux pump and therefore can overcome MDR. However, secondary effects of NMP other than efflux pump inhibition are rarely investigated. Here, using phenotypic assays, phenotypic microarray and transcriptomic assays we show that NMP creates membrane destabilization in MDR Klebsiella pneumoniae MGH 78578 strain. The NMP mediated membrane destabilization activity was measured using β-lactamase activity, membrane potential alteration studies, and transmission electron microscopy assays. Results from both β-lactamase and membrane potential alteration studies shows that both outer and inner membranes are destabilized in NMP exposed K. pneumoniae MGH 78578 cells. Phenotypic Microarray and RNA-seq were further used to elucidate the metabolic and transcriptional signals underpinning membrane destabilization. Membrane destabilization happens as early as 15 min post-NMP treatment. Our RNA-seq data shows that many genes involved in envelope stress response were differentially regulated in the NMP treated cells. Up-regulation of genes encoding the envelope stress response and repair systems show the distortion in membrane homeostasis during survival in an environment containing sub-inhibitory concentration of NMP. In addition, the lsr operon encoding the production of autoinducer-2 responsible for biofilm production was down-regulated resulting in reduced biofilm formation in NMP treated cells, a phenotype confirmed by crystal violet-based assays. We postulate that the early membrane disruption leads to destabilization of inner membrane potential, impairing ATP production and consequently resulting in efflux pump inhibition.
Keywords: 1-(1-naphthylmethyl)-piperazine; Klebsiella pneumoniae; NMP; chemosensitizers; membrane destabilization; secondary effects.
Figures


References
-
- Bina X. R., Philippart J. A., Bina J. E. (2009). Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J. Antimicrob. Chemother. 63 103–108. 10.1093/jac/dkn466 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

