Genotoxic stress increases cytoplasmic mitochondrial DNA editing by human APOBEC3 mutator enzymes at a single cell level
- PMID: 30816165
- PMCID: PMC6395610
- DOI: 10.1038/s41598-019-39245-8
Genotoxic stress increases cytoplasmic mitochondrial DNA editing by human APOBEC3 mutator enzymes at a single cell level
Abstract
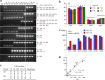
Human cells are stressed by numerous mechanisms that can lead to leakage of mitochondrial DNA (mtDNA) to the cytoplasm and ultimately apoptosis. This agonist DNA constitutes a danger to the cell and is counteracted by cytoplasmic DNases and APOBEC3 cytidine deamination of DNA. To investigate APOBEC3 editing of leaked mtDNA to the cytoplasm, we performed a PCR analysis of APOBEC3 edited cytoplasmic mtDNA (cymtDNA) at the single cell level for primary CD4+ T cells and the established P2 EBV blast cell line. Up to 17% of primary CD4+ T cells showed signs of APOBEC3 edited cymtDNA with ~50% of all mtDNA sequences showing signs of APOBEC3 editing - between 1500-5000 molecules. Although the P2 cell line showed a much lower frequency of stressed cells, the number of edited mtDNA molecules in such cells was of the same order. Addition of the genotoxic molecules, etoposide or actinomycin D increased the number of cells showing APOBEC3 edited cymtDNA to around 40%. These findings reveal a very dynamic image of the mitochondrial network, which changes considerably under stress. APOBEC3 deaminases are involved in the catabolism of mitochondrial DNA to circumvent chronic immune stimulation triggered by released mitochondrial DNA from damaged cells.
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Self-cytoplasmic DNA upregulates the mutator enzyme APOBEC3A leading to chromosomal DNA damage.Nucleic Acids Res. 2017 Apr 7;45(6):3231-3241. doi: 10.1093/nar/gkx001. Nucleic Acids Res. 2017. PMID: 28100701 Free PMC article.
-
Somatic hypermutation of human mitochondrial and nuclear DNA by APOBEC3 cytidine deaminases, a pathway for DNA catabolism.Proc Natl Acad Sci U S A. 2011 Mar 22;108(12):4858-63. doi: 10.1073/pnas.1009687108. Epub 2011 Mar 2. Proc Natl Acad Sci U S A. 2011. PMID: 21368204 Free PMC article.
-
APOBEC3A damages the cellular genome during DNA replication.Cell Cycle. 2016;15(7):998-1008. doi: 10.1080/15384101.2016.1152426. Cell Cycle. 2016. PMID: 26918916 Free PMC article.
-
Diverse functions for DNA and RNA editing in the immune system.RNA Biol. 2010 Mar-Apr;7(2):220-8. doi: 10.4161/rna.7.2.11344. Epub 2010 Mar 29. RNA Biol. 2010. PMID: 20220309 Review.
-
[Molecular evolution of physiologically functioning anti-retroviral APOBEC3 deaminases].Uirusu. 2012 Jun;62(1):27-38. doi: 10.2222/jsv.62.27. Uirusu. 2012. PMID: 23189822 Review. Japanese.
Cited by
-
Sustained high expression of multiple APOBEC3 cytidine deaminases in systemic lupus erythematosus.Sci Rep. 2021 Apr 12;11(1):7893. doi: 10.1038/s41598-021-87024-1. Sci Rep. 2021. PMID: 33846459 Free PMC article.
-
Mitochondrial DNA Release in Innate Immune Signaling.Annu Rev Biochem. 2023 Jun 20;92:299-332. doi: 10.1146/annurev-biochem-032620-104401. Epub 2023 Mar 31. Annu Rev Biochem. 2023. PMID: 37001140 Free PMC article. Review.
-
Clearing of Foreign Episomal DNA from Human Cells by CRISPRa-Mediated Activation of Cytidine Deaminases.Int J Mol Sci. 2020 Sep 18;21(18):6865. doi: 10.3390/ijms21186865. Int J Mol Sci. 2020. PMID: 32962129 Free PMC article.
-
Mouse APOBEC1 cytidine deaminase can induce somatic mutations in chromosomal DNA.BMC Genomics. 2019 Nov 14;20(1):858. doi: 10.1186/s12864-019-6216-x. BMC Genomics. 2019. PMID: 31726973 Free PMC article.
-
Critical contribution of mitochondria in the development of cardiomyopathy linked to desmin mutation.Stem Cell Res Ther. 2024 Jan 2;15(1):10. doi: 10.1186/s13287-023-03619-7. Stem Cell Res Ther. 2024. PMID: 38167524 Free PMC article.
References
-
- Langlois MA, Beale RC, Conticello SG, Neuberger MS. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res. 2005;33:1913–1923. doi: 10.1093/nar/gki343. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

