High-resolution atmospheric-pressure MALDI mass spectrometry imaging workflow for lipidomic analysis of late fetal mouse lungs
- PMID: 30816198
- PMCID: PMC6395778
- DOI: 10.1038/s41598-019-39452-3
High-resolution atmospheric-pressure MALDI mass spectrometry imaging workflow for lipidomic analysis of late fetal mouse lungs
Abstract
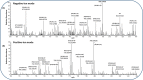
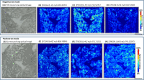
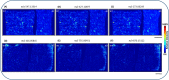
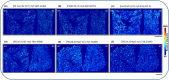
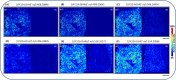
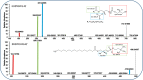
Mass spectrometry imaging (MSI) provides label-free, non-targeted molecular and spatial information of the biomolecules within tissue. Lipids play important roles in lung biology, e.g. as surfactant, preventing alveolar collapse during normal and forced respiration. Lipidomic characterization of late fetal mouse lungs at day 19 of gestation (E19) has not been performed yet. In this study we employed high-resolution atmospheric pressure scanning microprobe matrix-assisted laser desorption/ionization MSI for the lipidomic analysis of E19 mouse lungs. Molecular species of different lipid classes were imaged in E19 lung sections at high spatial and mass resolution in positive- and negative-ion mode. Lipid species were characterized based on accurate mass and on-tissue tandem mass spectrometry. In addition, a dedicated sample preparation protocol, homogenous deposition of matrices on tissue surfaces and data processing parameters were optimized for the comparison of signal intensities of lipids between different tissue sections of E19 lungs of wild type and Pex11β-knockout mice. Our study provides lipid information of E19 mouse lungs, optimized experimental and data processing strategies for the direct comparison of signal intensities of metabolites (lipids) among the tissue sections from MSI experiments. To best of our knowledge, this is the first MSI and lipidomic study of E19 mouse lungs.
Conflict of interest statement
B.S. is a consultant of TransMIT GmbH, Giessen, Germany. D.R.B. is a part-time employee of TransMIT GmbH, Giessen, Germany. The other authors declare no competing interests.
Figures



Similar articles
-
Lipid Coverage in Nanospray Desorption Electrospray Ionization Mass Spectrometry Imaging of Mouse Lung Tissues.Anal Chem. 2019 Sep 17;91(18):11629-11635. doi: 10.1021/acs.analchem.9b02045. Epub 2019 Aug 27. Anal Chem. 2019. PMID: 31412198 Free PMC article.
-
Enhanced capabilities for imaging gangliosides in murine brain with matrix-assisted laser desorption/ionization and desorption electrospray ionization mass spectrometry coupled to ion mobility separation.Methods. 2016 Jul 15;104:69-78. doi: 10.1016/j.ymeth.2016.02.014. Epub 2016 Feb 23. Methods. 2016. PMID: 26922843
-
Matrix-free mass spectrometry imaging of mouse brain tissue sections on silicon nanopost arrays.J Comp Neurol. 2019 Sep 1;527(13):2101-2121. doi: 10.1002/cne.24566. Epub 2018 Dec 5. J Comp Neurol. 2019. PMID: 30358893
-
Surface analysis of lipids by mass spectrometry: more than just imaging.Prog Lipid Res. 2013 Oct;52(4):329-53. doi: 10.1016/j.plipres.2013.04.005. Epub 2013 Apr 24. Prog Lipid Res. 2013. PMID: 23623802 Review.
-
Spatial Metabolite Profiling by Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Imaging.Adv Exp Med Biol. 2017;965:291-321. doi: 10.1007/978-3-319-47656-8_12. Adv Exp Med Biol. 2017. PMID: 28132185 Review.
Cited by
-
High-resolution imaging and identification of biomolecules using Nano-DESI coupled to ion mobility spectrometry.Anal Chim Acta. 2021 Nov 22;1186:339085. doi: 10.1016/j.aca.2021.339085. Epub 2021 Sep 21. Anal Chim Acta. 2021. PMID: 34756271 Free PMC article.
-
Lipid mediators of inhalation exposure-induced pulmonary toxicity and inflammation.Inhal Toxicol. 2024 Feb;36(2):57-74. doi: 10.1080/08958378.2024.2318389. Epub 2024 Feb 29. Inhal Toxicol. 2024. PMID: 38422051 Free PMC article. Review.
-
Comparative lipid profiling of murine and human atherosclerotic plaques using high-resolution MALDI MSI.Pflugers Arch. 2022 Feb;474(2):231-242. doi: 10.1007/s00424-021-02643-x. Epub 2021 Nov 19. Pflugers Arch. 2022. PMID: 34797426 Free PMC article.
-
Metabotype analysis of Mthfd1l-null mouse embryos using desorption electrospray ionization mass spectrometry imaging.Anal Bioanal Chem. 2021 May;413(13):3573-3582. doi: 10.1007/s00216-021-03308-5. Epub 2021 Apr 8. Anal Bioanal Chem. 2021. PMID: 33829277
-
Lipid Changes in the Peri-Implantation Period with Mass Spectrometry Imaging: A Systematic Review.Life (Basel). 2023 Jan 6;13(1):169. doi: 10.3390/life13010169. Life (Basel). 2023. PMID: 36676119 Free PMC article. Review.
References
-
- King, R. J. & Clements, J. A. In Comprehensive Physiology (John Wiley & Sons, Inc., 2011).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

