Impact of branching on the conformational heterogeneity of the lipopolysaccharide from Klebsiella pneumoniae: Implications for vaccine design
- PMID: 30818097
- PMCID: PMC6529184
- DOI: 10.1016/j.carres.2019.02.003
Impact of branching on the conformational heterogeneity of the lipopolysaccharide from Klebsiella pneumoniae: Implications for vaccine design
Abstract
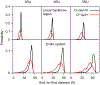
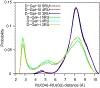
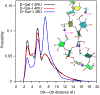
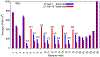
Resistance of Klebsiella pneumoniae (KP) to antibiotics has motivated the development of an efficacious KP human vaccine that would not be subject to antibiotic resistance. Klebsiella lipopolysaccharide (LPS) associated O polysaccharide (OPS) types have provoked broad interest as a vaccine antigen as there are only 4 that predominate worldwide (O1, O2a, O3, O5). Klebsiella O1 and O2 OPS are polygalactans that share a common D-Gal-I structure, for which a variant D-Gal-III was recently discovered. To understand the potential impact of this variability on antigenicity, a detailed molecular picture of the conformational differences associated with the addition of the D-Gal-III (1 → 4)-α-Galp branch is presented using enhanced-sampling molecular dynamics simulations. In D-Gal-I two major conformational states are observed while the presence of the 1 → 4 branch in D-Gal-III resulted in only a single dominant extended state. Stabilization of the more folded states in D-Gal-I is due to a O4-H⋯O2 hydrogen bond in the linear backbone that cannot occur in D-Gal-III as the O4 is in the Galp(1 → 4)Galp glycosidic linkage. The impact of branching in D-Gal-III also significantly decreases the accessibility of the monosaccharides in the linear backbone region of D-Gal-I, while the accessibility of the terminal D-Gal-II region of the OPS is not substantially altered. The present results suggest that a vaccine that targets both the D-Gal-I and D-Gal-III LPS can be developed by using D-Gal-III as the antigen combined with cross-reactivity experiments using the Gal-II polysaccharide to assure that this region of the LPS is the primary epitope of the antigen.
Keywords: Antibiotic resistance; Antigen; Antimicrobial resistance; CHARMM; Empirical force field; Lipopolysaccharide; Molecular dynamics simulations; Pneumonia.
Copyright © 2019 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Competing Financial Interests Statement: ADM Jr. is cofounder and CSO of SilcsBio LLC.
Figures







Similar articles
-
The structure of the core region of the lipopolysaccharide from Klebsiella pneumoniae O3. 3-deoxy-alpha-D-manno-octulosonic acid (alpha-Kdo) residue in the outer part of the core, a common structural element of Klebsiella pneumoniae O1, O2, O3, O4, O5, O8, and O12 lipopolysaccharides.Eur J Biochem. 2001 Mar;268(6):1722-9. doi: 10.1046/j.1432-1033.2001.02047.x. Eur J Biochem. 2001. PMID: 11248692
-
Klebsiella pneumoniae O1 and O2ac antigens provide prototypes for an unusual strategy for polysaccharide antigen diversification.J Biol Chem. 2019 Jul 12;294(28):10863-10876. doi: 10.1074/jbc.RA119.008969. Epub 2019 May 28. J Biol Chem. 2019. PMID: 31138653 Free PMC article.
-
Molecular basis for the structural diversity in serogroup O2-antigen polysaccharides in Klebsiella pneumoniae.J Biol Chem. 2018 Mar 30;293(13):4666-4679. doi: 10.1074/jbc.RA117.000646. Epub 2018 Feb 12. J Biol Chem. 2018. PMID: 29602878 Free PMC article.
-
Structural analysis of the core region of the lipopolysaccharides from eight serotypes of Klebsiella pneumoniae.Carbohydr Res. 2001 Oct 15;335(4):291-6. doi: 10.1016/s0008-6215(01)00216-6. Carbohydr Res. 2001. PMID: 11595223
-
Identification of d-Galactan-III As Part of the Lipopolysaccharide of Klebsiella pneumoniae Serotype O1.Front Microbiol. 2017 Apr 25;8:684. doi: 10.3389/fmicb.2017.00684. eCollection 2017. Front Microbiol. 2017. PMID: 28487676 Free PMC article.
Cited by
-
Klebsiella pneumoniae Lipopolysaccharide as a Vaccine Target and the Role of Antibodies in Protection from Disease.Vaccines (Basel). 2024 Oct 17;12(10):1177. doi: 10.3390/vaccines12101177. Vaccines (Basel). 2024. PMID: 39460343 Free PMC article. Review.
-
Fighting Against Bacterial Lipopolysaccharide-Caused Infections through Molecular Dynamics Simulations: A Review.J Chem Inf Model. 2021 Oct 25;61(10):4839-4851. doi: 10.1021/acs.jcim.1c00613. Epub 2021 Sep 24. J Chem Inf Model. 2021. PMID: 34559524 Free PMC article. Review.
-
O-antigen polysaccharides in Klebsiella pneumoniae: structures and molecular basis for antigenic diversity.Microbiol Mol Biol Rev. 2025 Jun 25;89(2):e0009023. doi: 10.1128/mmbr.00090-23. Epub 2025 Mar 21. Microbiol Mol Biol Rev. 2025. PMID: 40116577 Review.
-
Host Synthesized Carbohydrate Antigens on Viral Glycoproteins as "Achilles' Heel" of Viruses Contributing to Anti-Viral Immune Protection.Int J Mol Sci. 2020 Sep 13;21(18):6702. doi: 10.3390/ijms21186702. Int J Mol Sci. 2020. PMID: 32933166 Free PMC article. Review.
-
Modeling of pneumococcal serogroup 10 capsular polysaccharide molecular conformations provides insight into epitopes and observed cross-reactivity.Front Mol Biosci. 2022 Aug 8;9:961532. doi: 10.3389/fmolb.2022.961532. eCollection 2022. Front Mol Biosci. 2022. PMID: 36003080 Free PMC article.
References
-
- Vinogradov E, Frirdich E, MacLean LL, Perry MB, Petersen BO, Duus J, et al. Structures of lipopolysaccharides from Klebsiella pneumoniae: Elucidation of the structure of the linkage region between core and polysaccharide O chain and identification of the residues at the non-reducing termini of the O chains. J Biol Chem. 2002. 10.1074/jbc.M202683200 - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

