CyanoGate: A Modular Cloning Suite for Engineering Cyanobacteria Based on the Plant MoClo Syntax
- PMID: 30819783
- PMCID: PMC6501082
- DOI: 10.1104/pp.18.01401
CyanoGate: A Modular Cloning Suite for Engineering Cyanobacteria Based on the Plant MoClo Syntax
Abstract
Recent advances in synthetic biology research have been underpinned by an exponential increase in available genomic information and a proliferation of advanced DNA assembly tools. The adoption of plasmid vector assembly standards and parts libraries has greatly enhanced the reproducibility of research and the exchange of parts between different labs and biological systems. However, a standardized modular cloning (MoClo) system is not yet available for cyanobacteria, which lag behind other prokaryotes in synthetic biology despite their huge potential regarding biotechnological applications. By building on the assembly library and syntax of the Plant Golden Gate MoClo kit, we have developed a versatile system called CyanoGate that unites cyanobacteria with plant and algal systems. Here, we describe the generation of a suite of parts and acceptor vectors for making (1) marked/unmarked knock-outs or integrations using an integrative acceptor vector, and (2) transient multigene expression and repression systems using known and previously undescribed replicative vectors. We tested and compared the CyanoGate system in the established model cyanobacterium Synechocystis sp. PCC 6803 and the more recently described fast-growing strain Synechococcus elongatus UTEX 2973. The UTEX 2973 fast-growth phenotype was only evident under specific growth conditions; however, UTEX 2973 accumulated high levels of proteins with strong native or synthetic promoters. The system is publicly available and can be readily expanded to accommodate other standardized MoClo parts to accelerate the development of reliable synthetic biology tools for the cyanobacterial community.
© 2019 American Society of Plant Biologists. All Rights Reserved.
Figures



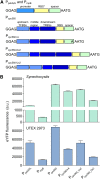


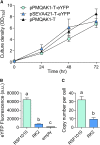
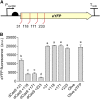
Comment in
-
A Bridge between Kingdoms: Introduction of a Golden Gate-Based Tool Kit for Cyanobacteria.Plant Physiol. 2019 May;180(1):10-11. doi: 10.1104/pp.19.00296. Plant Physiol. 2019. PMID: 31053673 Free PMC article. No abstract available.
References
-
- Abe K, Sakai Y, Nakashima S, Araki M, Yoshida W, Sode K, Ikebukuro K (2014) Design of riboregulators for control of cyanobacterial (Synechocystis) protein expression. Biotechnol Lett 36: 287–294 - PubMed
-
- Albers SC, Gallegos VA, Peebles CAM (2015) Engineering of genetic control tools in Synechocystis sp. PCC 6803 using rational design techniques. J Biotechnol 216: 36–46 - PubMed
-
- Andersson CR, Tsinoremas NF, Shelton J, Lebedeva NV, Yarrow J, Min H, Golden SS (2000) Application of bioluminescence to the study of circadian rhythms in cyanobacteria. Methods Enzymol 305: 527–542 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

