Auxin regulates endosperm cellularization in Arabidopsis
- PMID: 30819818
- PMCID: PMC6446538
- DOI: 10.1101/gad.316554.118
Auxin regulates endosperm cellularization in Arabidopsis
Abstract
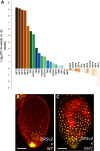
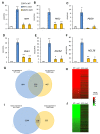
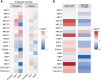
The endosperm is an ephemeral tissue that nourishes the developing embryo, similar to the placenta in mammals. In most angiosperms, endosperm development starts as a syncytium, in which nuclear divisions are not followed by cytokinesis. The timing of endosperm cellularization largely varies between species, and the event triggering this transition remains unknown. Here we show that increased auxin biosynthesis in the endosperm prevents its cellularization, leading to seed arrest. Auxin-overproducing seeds phenocopy paternal-excess triploid seeds derived from hybridizations of diploid maternal plants with tetraploid fathers. Concurrently, auxin-related genes are strongly overexpressed in triploid seeds, correlating with increased auxin activity. Reducing auxin biosynthesis and signaling reestablishes endosperm cellularization in triploid seeds and restores their viability, highlighting a causal role of increased auxin in preventing endosperm cellularization. We propose that auxin determines the time of endosperm cellularization, and thereby uncovered a central role of auxin in establishing hybridization barriers in plants.
Keywords: auxin; cellularization; endosperm; hybridization barrier; seed development; triploid block.
© 2019 Batista et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Belmonte MF, Kirkbride RC, Stone SL, Pelletier JM, Bui AQ, Yeung EC, Hashimoto M, Fei J, Harada CM, Munoz MD, et al. 2013. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc Natl Acad Sci 110: E435–E444. 10.1073/pnas.1222061110 - DOI - PMC - PubMed
-
- Boer DR, Freire-Rios A, van den Berg WAM, Saaki T, Manfield IW, Kepinski S, López-Vidrieo I, Franco-Zorrilla JM, de Vries SC, Solano R, et al. 2014. Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156: 577–589. 10.1016/j.cell.2013.12.027 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
