CRISPR-PIN: Modifying gene position in the nucleus via dCas9-mediated tethering
- PMID: 30820479
- PMCID: PMC6378893
- DOI: 10.1016/j.synbio.2019.02.001
CRISPR-PIN: Modifying gene position in the nucleus via dCas9-mediated tethering
Erratum in
-
Erratum regarding previously published articles.Synth Syst Biotechnol. 2020 Oct 14;5(4):330-331. doi: 10.1016/j.synbio.2020.10.001. eCollection 2020 Dec. Synth Syst Biotechnol. 2020. PMID: 33102827 Free PMC article.
Abstract
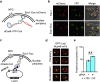
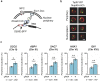
Spatial organization of DNA within the nucleus is important for controlling DNA replication and repair, genetic recombination, and gene expression. Here, we present CRISPR-PIN, a CRISPR/dCas9-based tool that allows control of gene Position in the Nucleus for the yeast Saccharomyces cerevisiae. This approach utilizes a cohesin-dockerin interaction between dCas9 and a perinuclear protein. In doing so, we demonstrate that a single gRNA can enable programmable interaction of nuclear DNA with the nuclear periphery. We demonstrate the utility of this approach for two applications: the controlled segregation of an acentric plasmid and the re-localization of five endogenous loci. In both cases, we obtain results on par with prior reports using traditional, more cumbersome genetic systems. Thus, CRISPR-PIN offers the opportunity for future studies of chromosome biology and gene localization.
Keywords: CRISPR; Chromosome biology; Chromosome organization; Gene positioning; Synthetic biology.
Figures


References
-
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. - PubMed
-
- Baudat F., Imai Y., de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14:794–806. - PubMed
-
- de Juan D., Pazos F., Valencia A. Emerging methods in protein co-evolution. Nat Rev Genet. 2013;14:249–261. - PubMed
-
- Marsh J.A., Teichmann S.A. Structure, dynamics, assembly, and evolution of protein complexes. Annu Rev Biochem. 2015;84:551–575. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

