CRISPRs for Strain Tracking and Their Application to Microbiota Transplantation Data Analysis
- PMID: 30820491
- PMCID: PMC6390457
- DOI: 10.1089/crispr.2018.0046
CRISPRs for Strain Tracking and Their Application to Microbiota Transplantation Data Analysis
Abstract
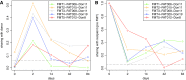
CRISPR-Cas systems are adaptive immune systems naturally found in bacteria and archaea. Prokaryotes use these immune systems to defend against invaders, which include phages, plasmids, and other mobile genetic elements. Relying on the integration of spacers derived from invader sequences (protospacers) into CRISPR loci (forming spacers flanked by repeats), CRISPR-Cas systems are able to store the memory of past immunological encounters. While CRISPR-Cas systems have evolved in response to invading mobile genetic elements, invaders have also developed mechanisms to avoid detection. As a result of an arms race between CRISPR-Cas systems and their targets, CRISPR arrays typically undergo rapid turnover of spacers through the acquisition and loss events. Additionally, microbiomes of different individuals rarely share spacers. Here, we present a computational pipeline, CRISPRtrack, for strain tracking based on CRISPR spacer content, and we applied it to fecal transplantation microbiome data to study the retention of donor strains in recipients. Our results demonstrate the potential use of CRISPRs as a simple yet effective tool for donor-strain tracking in fecal transplantation and as a general purpose tool for quantifying microbiome similarity.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
