Hormone therapy use and breast tissue DNA methylation: analysis of epigenome wide data from the normal breast study
- PMID: 30821641
- PMCID: PMC6557608
- DOI: 10.1080/15592294.2019.1580111
Hormone therapy use and breast tissue DNA methylation: analysis of epigenome wide data from the normal breast study
Abstract
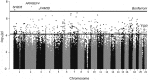
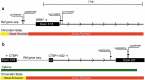
Hormone therapy (HT) is associated with increased risk of breast cancer, strongly dependent on type, duration, and recency of use. HT use could affect cancer risk by changing breast tissue transcriptional programs. We hypothesize that these changes are preceded by changes in DNA methylation. To explore this hypothesis we used histologically normal-appearing breast tissue from the Normal Breast Study (NBS). DNA methylation β-values were obtained using the Illumina HumanMethylation 450 BeadChips for 90 samples including all NBS-participants who used HT within 5 y before surgery. Data were analyzed using the reference-free cell mixture method. Cancer Genome Atlas (TCGA) mRNA-Seq data were used to assess correlation between DNA methylation and gene expression. We identified 527 CpG sites in 403 genes that were associated with ever using HT at genome wide significance (FDR q < 0.05), of these, 68 sites were also significantly associated with duration of use or recency of use. Twelve sites reached significance in all analyses one of which was cg01382688 in ARHGEF4 (p < 1.2x10-7). Mutations in ARHGEF4 have been reported in breast tumors, but this is the first report of possible breast cancer-related DNA methylation changes. In addition, 22 genes included more than one significant CpG site and a majority of these sites were significantly correlated with gene expression. Although based on small numbers, these findings support the hypothesis that HT is associated with epigenetic alterations in breast tissue, and identifies genes with altered DNA methylation states which could be linked to breast cancer development.
Keywords: DNA methylation; Hormone therapy; epigenetics; epigenome wide association; normal breast tissue.
Figures
Similar articles
-
Epigenome-wide association study for lifetime estrogen exposure identifies an epigenetic signature associated with breast cancer risk.Clin Epigenetics. 2019 Apr 30;11(1):66. doi: 10.1186/s13148-019-0664-7. Clin Epigenetics. 2019. PMID: 31039828 Free PMC article.
-
Normal breast tissue DNA methylation differences at regulatory elements are associated with the cancer risk factor age.Breast Cancer Res. 2017 Jul 10;19(1):81. doi: 10.1186/s13058-017-0873-y. Breast Cancer Res. 2017. PMID: 28693600 Free PMC article.
-
DNA methylation in childhood asthma: an epigenome-wide meta-analysis.Lancet Respir Med. 2018 May;6(5):379-388. doi: 10.1016/S2213-2600(18)30052-3. Epub 2018 Feb 26. Lancet Respir Med. 2018. PMID: 29496485
-
Tools and Strategies for Analysis of Genome-Wide and Gene-Specific DNA Methylation Patterns.Methods Mol Biol. 2017;1537:249-277. doi: 10.1007/978-1-4939-6685-1_15. Methods Mol Biol. 2017. PMID: 27924599 Review.
-
Epigenome-wide DNA methylation and risk of breast cancer: a systematic review.BMC Cancer. 2020 Oct 31;20(1):1048. doi: 10.1186/s12885-020-07543-4. BMC Cancer. 2020. PMID: 33129307 Free PMC article.
Cited by
-
Associations of breast cancer related exposures and gene expression profiles in normal breast tissue-The Norwegian Women and Cancer normal breast tissue study.Cancer Rep (Hoboken). 2023 Apr;6(4):e1777. doi: 10.1002/cnr2.1777. Epub 2023 Jan 8. Cancer Rep (Hoboken). 2023. PMID: 36617746 Free PMC article.
-
Ten Years of EWAS.Adv Sci (Weinh). 2021 Oct;8(20):e2100727. doi: 10.1002/advs.202100727. Epub 2021 Aug 11. Adv Sci (Weinh). 2021. PMID: 34382344 Free PMC article. Review.
References
-
- Zhu L, Jiang X, Sun Y, et al. Effect of hormone therapy on the risk of bone fractures: a systematic review and meta-analysis of randomized controlled trials. Menopause. 2016;23(4):461. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
