Oxidant-Mediated Protein Amino Acid Conversion
- PMID: 30823521
- PMCID: PMC6406366
- DOI: 10.3390/antiox8020050
Oxidant-Mediated Protein Amino Acid Conversion
Abstract
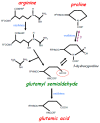
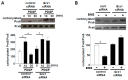
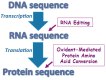
Biological oxidation plays important roles in the pathogenesis of various diseases and aging. Carbonylation is one mode of protein oxidation. It has been reported that amino acids that are susceptible to carbonylation are arginine (Arg), proline (Pro), lysine, and threonine residues. The carbonylation product of both Arg and Pro residues is glutamyl semialdehyde. While chemically the oxidation reactions of neither Pro to glutamyl semialdehyde nor Arg to glutamyl semialdehyde are reversible, experimental results from our laboratory suggest that the biological system may drive the reduction of glutamyl semialdehyde to Pro in the protein structure. Further, glutamyl semialdehyde can be oxidized to become glutamic acid (Glu). Therefore, I hypothesize that biological oxidation post-translationally converts Arg to Pro, Arg to Glu, and Pro to Glu within the protein structure. Our mass spectrometry experiments provided evidence that, in human cells, 5⁻10% of peroxiredoxin 6 protein molecules have Pro-45 replaced by Glu. This concept of protein amino acid conversion challenges the dogma that amino acid sequences are strictly defined by nucleic acid sequences. I propose that, in the biological system, amino acid replacements can occur post-translationally through redox regulation, and protein molecules with non-DNA coding sequences confer functions.
Keywords: amino acid; carbonylation; oxidation; protein; reactive oxygen species; redox.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Halliwell B., Gutteridge J. Free Radicals in Biology and Medicine. Oxford University Press; Oxford, UK: 2007.
-
- Amici A., Levine R.L., Tsai L., Stadtman E.R. Conversion of amino acid residues in proteins and amino acid homopolymers to carbonyl derivatives by metal-catalyzed oxidation reactions. J. Biol. Chem. 1989;264:3341–3346. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

