Transcriptomic Analysis for Indica and Japonica Rice Varieties under Aluminum Toxicity
- PMID: 30823582
- PMCID: PMC6412857
- DOI: 10.3390/ijms20040997
Transcriptomic Analysis for Indica and Japonica Rice Varieties under Aluminum Toxicity
Abstract
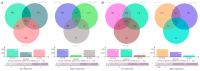
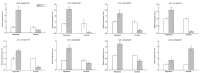
Aluminum (Al) at high concentrations inhibits root growth, damage root systems, and causes significant reductions in rice yields. Indica and Japonica rice have been cultivated in distinctly different ecological environments with different soil acidity levels; thus, they might have different mechanisms of Al-tolerance. In the present study, transcriptomic analysis in the root apex for Al-tolerance in the seedling stage was carried out within Al-tolerant and -sensitive varieties belonging to different subpopulations (i.e., Indica, Japonica, and mixed). We found that there were significant differences between the gene expression patterns of Indica Al-tolerant and Japonica Al-tolerant varieties, while the gene expression patterns of the Al-tolerant varieties in the mixed subgroup, which was inclined to Japonica, were similar to the Al-tolerant varieties in Japonica. Moreover, after further GO (gene ontology) and KEGG (Kyoto Encyclopedia of Genes and Genomes) analyses of the transcriptomic data, we found that eight pathways, i.e., "Terpenoid backbone biosynthesis", "Ribosome", "Amino sugar and nucleotide sugar metabolism", "Plant hormone signal transduction", "TCA cycle", "Synthesis and degradation of ketone bodies", and "Butanoate metabolism" were found uniquely for Indica Al-tolerant varieties, while only one pathway (i.e., "Sulfur metabolism") was found uniquely for Japonica Al-tolerant varieties. For Al-sensitive varieties, one identical pathway was found, both in Indica and Japonica. Three pathways were found uniquely in "Starch and sucrose metabolism", "Metabolic pathway", and "Amino sugar and nucleotide sugar metabolism".
Keywords: Indica; Japonica; aluminum toxicity; transcriptomic analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Vonuexkull H.R., Mutert E. Global extent, development and economic impact of acid soils. Plant Soil. 1995;171:1–15. doi: 10.1007/BF00009558. - DOI
-
- Liu J., Luo X., Shaff J., Liang C., Jia X., Li Z., Magalhaes J., Kochian L.V. A promoter-swap strategy between the AtALMT and AtMATE genes increased Arabidopsis aluminum resistance and improved carbon-use efficiency for aluminum resistance. Plant J. 2012;71:327–337. doi: 10.1111/j.1365-313X.2012.04994.x. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

