A signalling cascade involving receptor-activated phospholipase A2, glycerophosphoinositol 4-phosphate, Shp1 and Src in the activation of cell motility
- PMID: 30823936
- PMCID: PMC6396489
- DOI: 10.1186/s12964-019-0329-3
A signalling cascade involving receptor-activated phospholipase A2, glycerophosphoinositol 4-phosphate, Shp1 and Src in the activation of cell motility
Abstract
Background: Shp1, a tyrosine-phosphatase-1 containing the Src-homology 2 (SH2) domain, is involved in inflammatory and immune reactions, where it regulates diverse signalling pathways, usually by limiting cell responses through dephosphorylation of target molecules. Moreover, Shp1 regulates actin dynamics. One Shp1 target is Src, which controls many cellular functions including actin dynamics. Src has been previously shown to be activated by a signalling cascade initiated by the cytosolic-phospholipase A2 (cPLA2) metabolite glycerophosphoinositol 4-phosphate (GroPIns4P), which enhances actin polymerisation and motility. While the signalling cascade downstream Src has been fully defined, the mechanism by which GroPIns4P activates Src remains unknown.
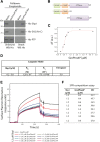
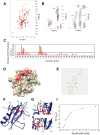
Methods: Affinity chromatography, mass spectrometry and co-immunoprecipitation studies were employed to identify the GroPIns4P-interactors; among these Shp1 was selected for further analysis. The specific Shp1 residues interacting with GroPIns4P were revealed by NMR and validated by site-directed mutagenesis and biophysical methods such as circular dichroism, isothermal calorimetry, fluorescence spectroscopy, surface plasmon resonance and computational modelling. Morphological and motility assays were performed in NIH3T3 fibroblasts.
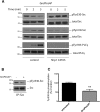
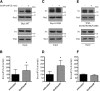
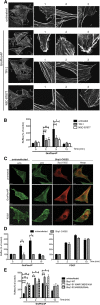
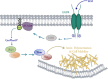
Results: We find that Shp1 is the direct cellular target of GroPIns4P. GroPIns4P directly binds to the Shp1-SH2 domain region (with the crucial residues being Ser 118, Arg 138 and Ser 140) and thereby promotes the association between Shp1 and Src, and the dephosphorylation of the Src-inhibitory phosphotyrosine in position 530, resulting in Src activation. As a consequence, fibroblast cells exposed to GroPIns4P show significantly enhanced wound healing capability, indicating that GroPIns4P has a stimulatory role to activate fibroblast migration. GroPIns4P is produced by cPLA2 upon stimulation by diverse receptors, including the EGF receptor. Indeed, endogenously-produced GroPIns4P was shown to mediate the EGF-induced cell motility.
Conclusions: This study identifies a so-far undescribed mechanism of Shp1/Src modulation that promotes cell motility and that is dependent on the cPLA2 metabolite GroPIns4P. We show that GroPIns4P is required for EGF-induced fibroblast migration and that it is part of a cPLA2/GroPIns4P/Shp1/Src cascade that might have broad implications for studies of immune-inflammatory response and cancer.
Keywords: Actin polymerisation; Cell motility; EGF; Glycerophosphoinositols; Membrane ruffles; Phosphoinositides; SH2 domain; Shp1.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Valitutti S, Cucchi P, Colletta G, Di Filippo C, Corda D. Transformation by the k-ras oncogene correlates with increases in phospholipase A2 activity, glycerophosphoinositol production and phosphoinositide synthesis in thyroid cells. Cell Signal. 1991;3:321–332. doi: 10.1016/0898-6568(91)90061-X. - DOI - PubMed
-
- Mariggio S, Sebastia J, Filippi BM, Iurisci C, Volonte C, Amadio S, De Falco V, Santoro M, Corda D. A novel pathway of cell growth regulation mediated by a PLA2alpha-derived phosphoinositide metabolite. The FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2006;20:2567–2569. doi: 10.1096/fj.05-5397fje. - DOI - PubMed
-
- Corda D, Falasca M. Glycerophosphoinositols as potential markers of ras-induced transformation and novel second messengers. Anticancer Res. 1996;16:1341–1350. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

